Detailed information of T6SS
Note: only experimentally validated proteins are displayed in the graph.
Summary
| T6SS ID | T6SS00016 |
| T6SS type | Type i5 |
| Strain | Agrobacterium fabrum str. C58 |
| Replicon (RefSeq) | chromosome linear (NC_003063) |
| Location | 1460756..1488734 |
| Function | anti-eukaryotic; anti-bacterial; effector translocation |
| Comment | Deletion of hcp results in reduced tumorigenesis on potato tuber slices; Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta; the Tde DNase toxins may be pivotal antibacterial toxins that A. tumefaciens uses against competitors during in planta colonization. |
| References | [1] Ma LS et al (2012) IcmF family protein TssM exhibits ATPase activity and energizes type VI secretion. J Biol Chem. 287(19):15610-21. [PudMed:22393043] [2] Lin JS et al (2013) Systematic Dissection of the Agrobacterium Type VI Secretion System Reveals Machinery and Secreted Components for Subcomplex Formation. PLoS One. 8(7):e67647. [PudMed:23861778] [3] Wu HY et al (2008) Secretome analysis uncovers an Hcp-family protein secreted via a type VI secretion system in Agrobacterium tumefaciens. J Bacteriol. 190(8):2841-50. [PudMed:18263727] [4] Ma LS et al (2009) An IcmF family protein, ImpLM, is an integral inner membrane protein interacting with ImpKL, and its walker a motif is required for type VI secretion system-mediated Hcp secretion in Agrobacterium tumefaciens. J Bacteriol. 191(13):4316-29. [PudMed:19395482] [5] Wu CF et al (2012) Acid-Induced Type VI Secretion System Is Regulated by ExoR-ChvG/ChvI Signaling Cascade in Agrobacterium tumefaciens. PLoS Pathog. 8(9):e1002938. [PudMed:23028331] [6] Ma LS et al (2014) Agrobacterium tumefaciens Deploys a Superfamily of Type VI Secretion DNase Effectors as Weapons for Interbacterial Competition In Planta. Cell Host Microbe. 16(1):94-104. [PudMed:24981331] [7] Heckel BC et al (2014) Agrobacterium tumefaciens ExoR Controls Acid Response Genes and Impacts Exopolysaccharide Synthesis, Horizontal Gene Transfer and Virulence Gene Expression. J Bacteriol. 196(18):3221-33. [PudMed:24982308] [8] Lin JS et al (2014) Fha Interaction with Phosphothreonine of TssL Activates Type VI Secretion in Agrobacterium tumefaciens. PLoS Pathog. 10(3):e1003991. [PudMed:24626341] |
T6SS components and genome coordinates
| Component | FHA | PAAR | PpkA | TagF-PppA | TagJ | TssA | TssB | TssC | TssD | TssE | TssF | TssG | TssH | TssI | TssK | TssL | TssM |
| Number | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 |
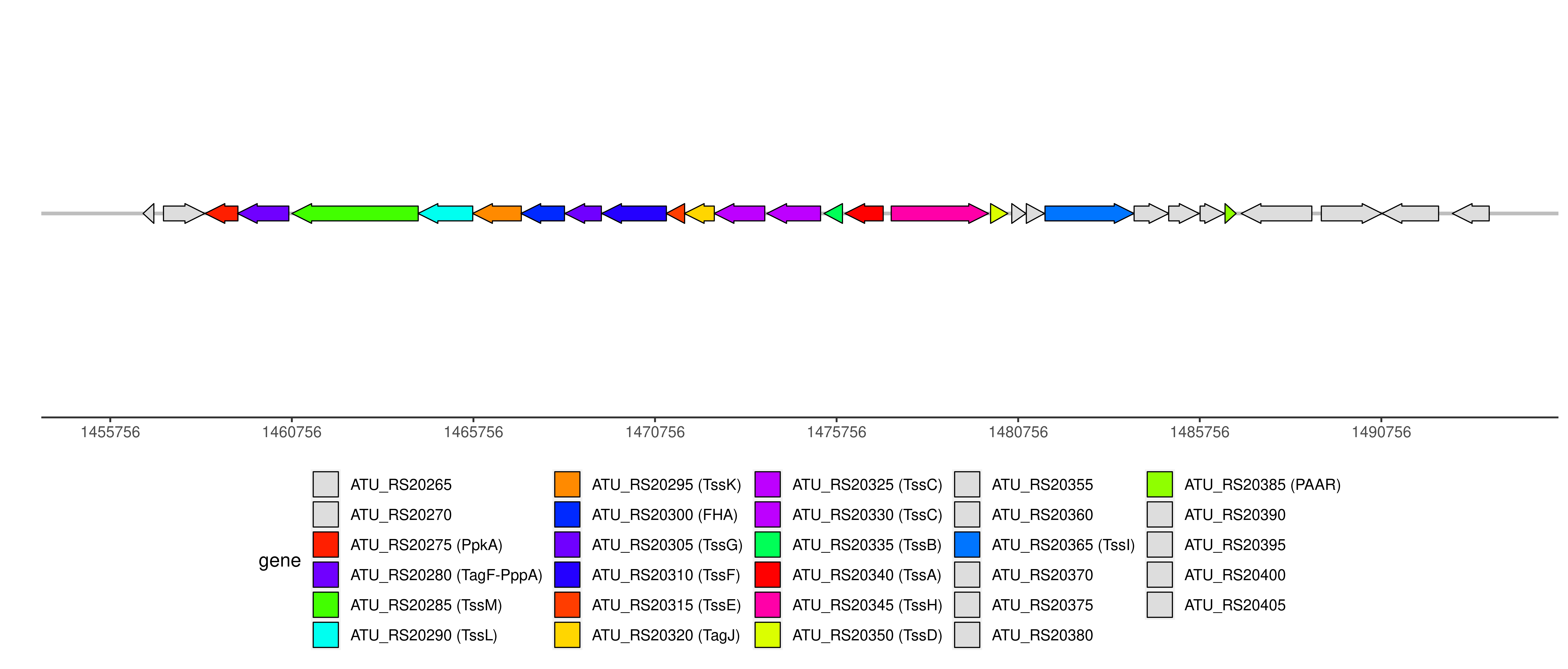
| Component ID | Component | Locus tag (Gene) | Coordinate | Strand | Size (bp) | NCBI ID | Product |
|---|---|---|---|---|---|---|---|
| ATU_RS20265 | 1456660..1456956 | - | 297 | WP_006315910.1 | PRC-barrel domain-containing protein | ||
| ATU_RS20270 | 1457221..1458357 | + | 1137 | WP_035257912.1 | polysaccharide export protein | ||
| T6CP012426 | PpkA | ATU_RS20275 | 1458366..1459271 | - | 906 | WP_035257928.1 | protein kinase |
| T6CP012427 | TagF-PppA | ATU_RS20280 (tagF) | 1459258..1460673 | - | 1416 | WP_010973754.1 | type VI secretion system-associated protein TagF |
| T6CP000114 | TssM | ATU_RS20285 (tssM) | 1460756..1464235 | - | 3480 | WP_010973755.1 | type VI secretion system membrane subunit TssM |
| T6CP000115 | TssL | ATU_RS20290 (tssL) | 1464232..1465737 | - | 1506 | WP_010973756.1 | type VI secretion system protein TssL, long form |
| T6CP000116 | TssK | ATU_RS20295 (tssK) | 1465734..1467074 | - | 1341 | WP_010973757.1 | type VI secretion system baseplate subunit TssK |
| T6CP011404 | FHA | ATU_RS20300 | 1467064..1468263 | - | 1200 | WP_010973758.1 | type VI secretion system-associated FHA domain protein |
| T6CP000117 | TssG | ATU_RS20305 (tssG) | 1468274..1469278 | - | 1005 | WP_010973759.1 | type VI secretion system baseplate subunit TssG |
| T6CP000118 | TssF | ATU_RS20310 (tssF) | 1469288..1471069 | - | 1782 | WP_010973760.1 | type VI secretion system baseplate subunit TssF |
| T6CP008386 | TssE | ATU_RS20315 (tssE) | 1471062..1471571 | - | 510 | WP_006315898.1 | type VI secretion system baseplate subunit TssE |
| T6CP012697 | TagJ | ATU_RS20320 | 1471564..1472388 | - | 825 | WP_006315897.1 | hypothetical protein |
| T6CP000119 | TssC | ATU_RS20325 (tssC) | 1472385..1473779 | - | 1395 | WP_006315896.1 | type VI secretion system contractile sheath large subunit |
| T6CP000120 | TssC | ATU_RS20330 (tssC) | 1473834..1475315 | - | 1482 | WP_006315895.1 | type VI secretion system contractile sheath large subunit |
| T6CP000121 | TssB | ATU_RS20335 (tssB) | 1475409..1475918 | - | 510 | WP_010973761.1 | type VI secretion system contractile sheath small subunit |
| T6CP009351 | TssA | ATU_RS20340 (tssA) | 1475984..1477039 | - | 1056 | WP_010973762.1 | type VI secretion system protein TssA |
| T6CP000122 | TssH | ATU_RS20345 (tssH) | 1477260..1479938 | + | 2679 | WP_010973763.1 | type VI secretion system ATPase TssH |
| T6CP000123 | TssD | ATU_RS20350 | 1479996..1480472 | + | 477 | WP_003504561.1 | type VI secretion system tube protein Hcp |
| ATU_RS20355 | 1480579..1480968 | + | 390 | WP_010973764.1 | hypothetical protein | ||
| ATU_RS20360 | 1480980..1481480 | + | 501 | WP_006315890.1 | type VI secretion system amidase effector protein Tae4 | ||
| T6CP000124 | TssI | ATU_RS20365 | 1481495..1483945 | + | 2451 | WP_010973765.1 | type VI secretion system tip protein VgrG |
| ATU_RS20370 | 1483948..1484904 | + | 957 | WP_010973766.1 | DUF4123 domain-containing protein | ||
| ATU_RS20375 | 1484901..1485737 | + | 837 | WP_010973767.1 | hypothetical protein | ||
| ATU_RS20380 | 1485765..1486439 | + | 675 | WP_010973768.1 | DUF1851 domain-containing protein | ||
| T6CP011354 | PAAR | ATU_RS20385 | 1486452..1486757 | + | 306 | WP_010973769.1 | PAAR domain-containing protein |
| ATU_RS20390 | 1486896..1488842 | - | 1947 | WP_035257634.1 | bifunctional diguanylate cyclase/phosphodiesterase | ||
| ATU_RS20395 | 1489103..1490773 | + | 1671 | WP_010973771.1 | metallophosphoesterase | ||
| ATU_RS20400 | 1490777..1492333 | - | 1557 | WP_010973772.1 | TerC family protein | ||
| ATU_RS20405 | 1492710..1493726 | - | 1017 | WP_010973773.1 | DUF1402 family protein |
Download FASTA format files
Note: in the 'Target' column, ↓ means the target is positively regulated by the regulator, while ↑ indicates negatively regulation.
| Effector ID | Locus tag (Gene) | Classification | Cognate immunity protein | Organism | Replicon (Accession) | Coordinates (Strand) | NCBI ID |
|---|---|---|---|---|---|---|---|
| EFF00996 |
Atu4347 (Tae4) | Tae | IMU00053 |
Agrobacterium fabrum str. C58 | chromosome (NC_003063) | 1480980..1481480 (+) | 159186127 |
| EFF01308 |
Atu4350 (Tde1) | Tde | - | Agrobacterium fabrum str. C58 | chromosome (NC_003063) | 1484901..1485737 (+) | 15890633 |
| EFF01307 |
Atu3640 (Tde2) | Tde | IMU00054 |
Agrobacterium fabrum str. C58 | chromosome (NC_003063) | 692241..693821 (-) | 15891300 |
| EFF00024 |
ATU_RS20350 | - | - | Agrobacterium fabrum str. C58 | chromosome (NC_003063) | 1479996..1480472 (+) | 15890638 |
| Immunity protein ID | Locus tag (Gene) | Cognate effector | Organism | Replicon (Accession) | Coordinates (Strand) | NCBI ID |
|---|---|---|---|---|---|---|
| IMU00053 |
Atu4351 (Tdi1) | EFF00996 |
Agrobacterium fabrum str. C58 | chromosome (NC_003063) | 1485765..1486439 (+) | 15890632 |
| IMU00054 |
Atu3639 (Tdi2) | EFF01307 |
Agrobacterium fabrum str. C58 | chromosome (NC_003063) | 691507..692238 (-) | 159185838 |
| IMU00192 |
Atu4346 (Tai4) | EFF00996 |
Agrobacterium fabrum str. C58 | chromosome (NC_003063) | 1480579..1480968 (+) | 159186126 |
| Accessory protein ID | Locus tag (Gene) | Type | Organism | Replicon (Accession) | Coordinates (Strand) | NCBI ID | Related structure protein | Related effector |
|---|---|---|---|---|---|---|---|---|
| ACP00010 |
Atu4349 (tap-1) | adaptor | Agrobacterium fabrum str. C58 | chromosome (NC_003063) | 1483948..1484904 (+) | 737273750 | Atu4348 (VgrG1) | Atu4350 (Tde1) |
| ACP00011 |
Atu3641 | adaptor | Agrobacterium fabrum str. C58 | chromosome (NC_003063) | 693818..694927 (-) | 499279473 | Atu3642 (VgrG2) | Atu3640 (Tde2) |