Introduction
Type VI secretion system (T6SS), a class of sophisticated protein delivering apparatus, usually composed of ~13 core components with or without accessory proteins in different species, and involved in mediating antagonistic or synergistic communications between bacteria and/or bacteria and eukaryotes. This apparatus has been found to be widely distributed among Gram-negative bacterial species and divided into four classes: i, ii, iii, and iv. Type i T6SS is the most classic and widely studied, and is further subclassed: i1, i2, i3, i4a, i4b, i5. To date, only a few datasets of T6SS core components are available, such as KEGG and MicrobeWiki. Two comprehensive databases EffectiveDB and BastionHub are mainly focus on T6SS effectors (T6SEs) and effectors of several secretion systems in bacteria, lacking specific and integrated full-scale data of T6SS and T6SS-related proteins such as T6SS regulator (T6SR), T6SS accessory protein (T6SA), T6SS immunity protein (T6SI), and recipient susceptibility factor. Here, we have organized information derived from the reported literature of T6SS gene cluster, T6SR, T6SA, recipient susceptibility factor, as well as T6SE, T6SI and new structure component protein, together with several tools for classification and prediction of T6SS and T6SS-related proteins as the web-based resource SecReT6 (Type VI Secretion system Resource) version 3.
SecReT6: a web-based resource for type VI secretion systems found in bacteria
The open-access database SecReT6 offers a unique, readily explorable archive of known and putative T6SSs and T6SS-related proteins including effectors, immunity proteins, regulators, accessory proteins and recipient susceptibility factors in bacteria via friendly and convenient interfaces.
A broad range of tools, including T6SS gene cluster and T6SS-related protein prediction, classification, comparative analyses, similarity searches, phylogenetic analysis, and T6SS-dependent killing risk estimation tools are readily accessible via SecReT6.
SecReT6 v3.0 released on Nov. 15th, 2021.
SecReT6 v3.0 currently contains 225 experimental validated T6SSs, 330 experimentally validated effectors, 156 experimentally validated cognate immunity proteins. 62 recipient susceptibility factor protein, 370 T6SS regulators, 49 T6SS accessory proteins are also archived in the database.
SecReT6 v3.0 has been refactored to exhibit via friendly and convenient web interfaces and data entrances in “Browse” and “Statistics” modules. The database integrated new tools to serve multiple functions including T6SS classification, T6SS-related protein prediction, T6SS prediction, T6SS-dependent killing risk estimation.
In addition, SecReT6 v3.0 also recorded computationally predicted T6SSs and their associated proteins in the “Download” module, including 17,223 T6SSs, 74,712 T6SEs, and 37805 T6SIs detected in 26,573 bacterial complete genomes currently available in NCBI.
SecReT6 v2.0 released on Jun. 28th, 2014.
SecReT6 v2.0 currently contains data on 10,891 core T6SS components belonging to 873 T6SSs found in 478 bacterial strains representing 221 species,
as well as a collection of over six hundred associated references.
Also collated and archived were 1,277 diverse candidate secreted effectors which were experimentally demonstrated and/or predicted to
be delivered by T6SSs into target eukaryotic and/or prokaryote cells as well as 174 immunity proteins.
SecReT6 v1.0 released on Aug. 15th, 2013.
SecReT6 v1.0 contains data on 9,664 core T6SS components mapping to 782 T6SSs found in 431 bacterial strains representing 216 species,
as well as a collection of over four hundred directly relevant references.
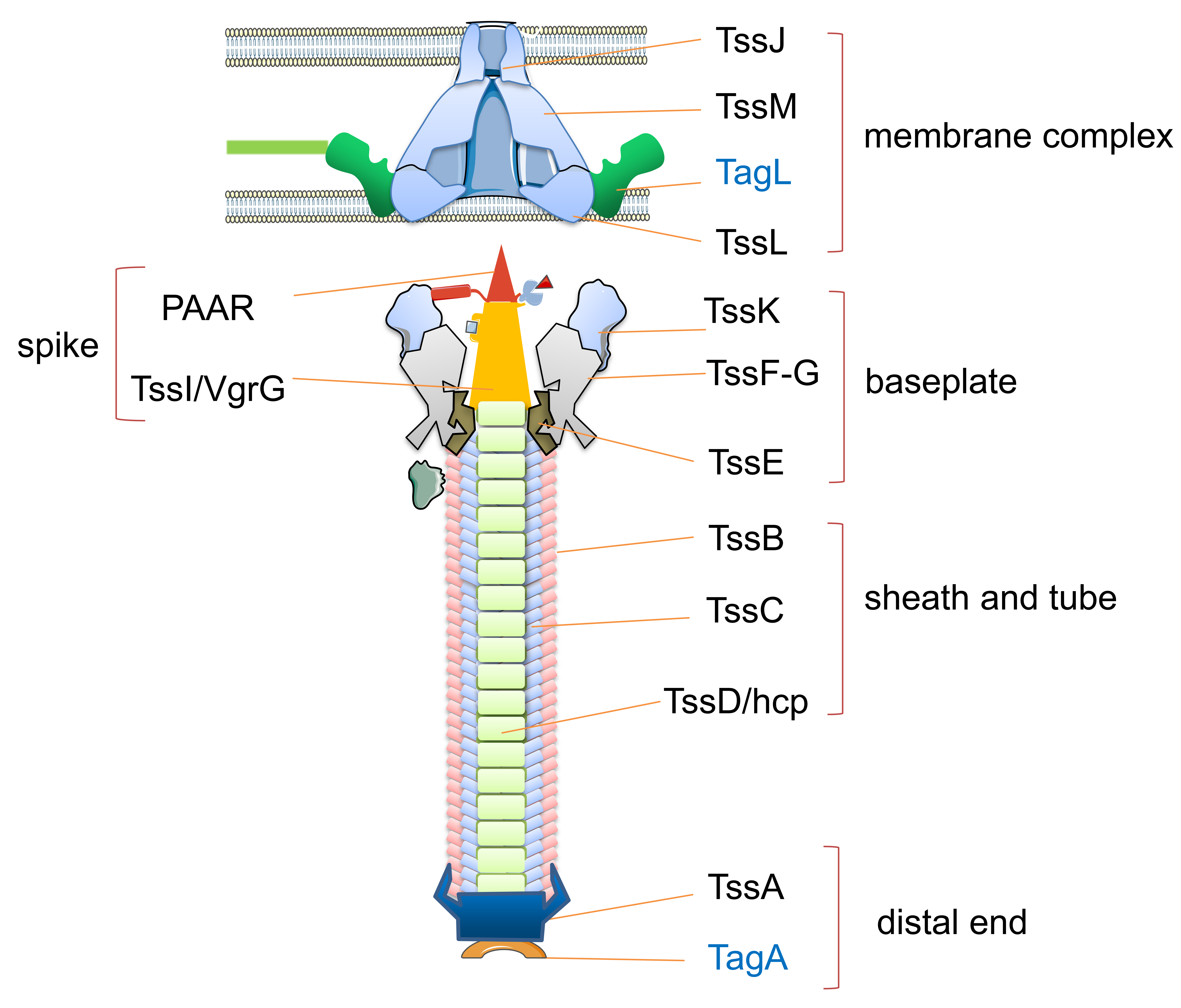
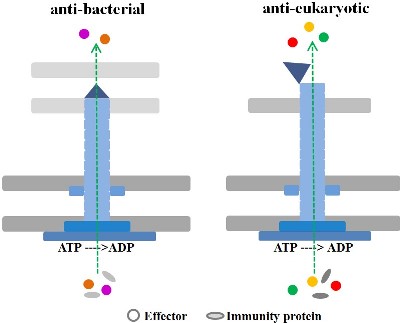
Overview of the classic T6SS assembly mode
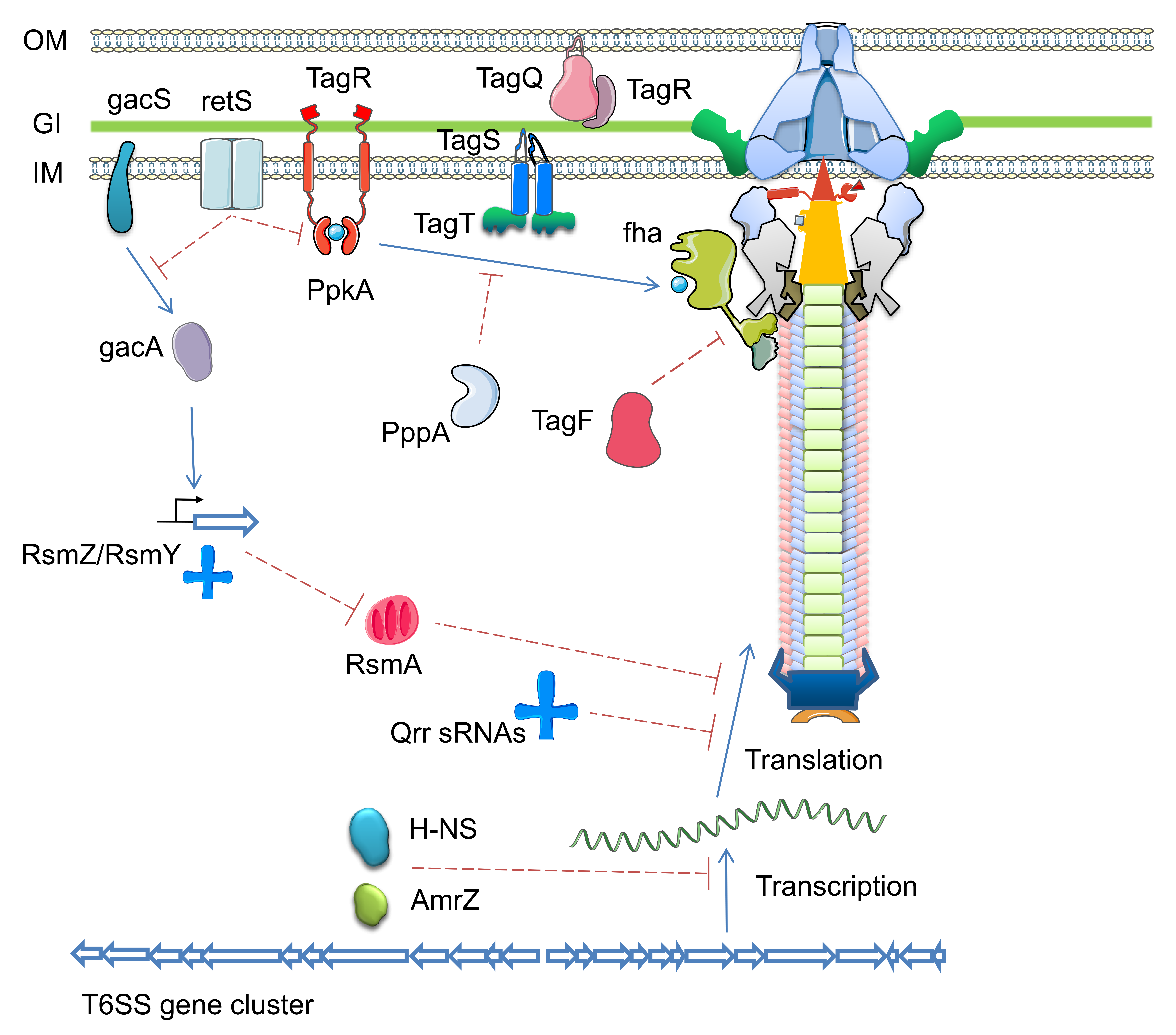
Overview of the T6SS regulatory mechanism in Pseudomonas aeruginosa
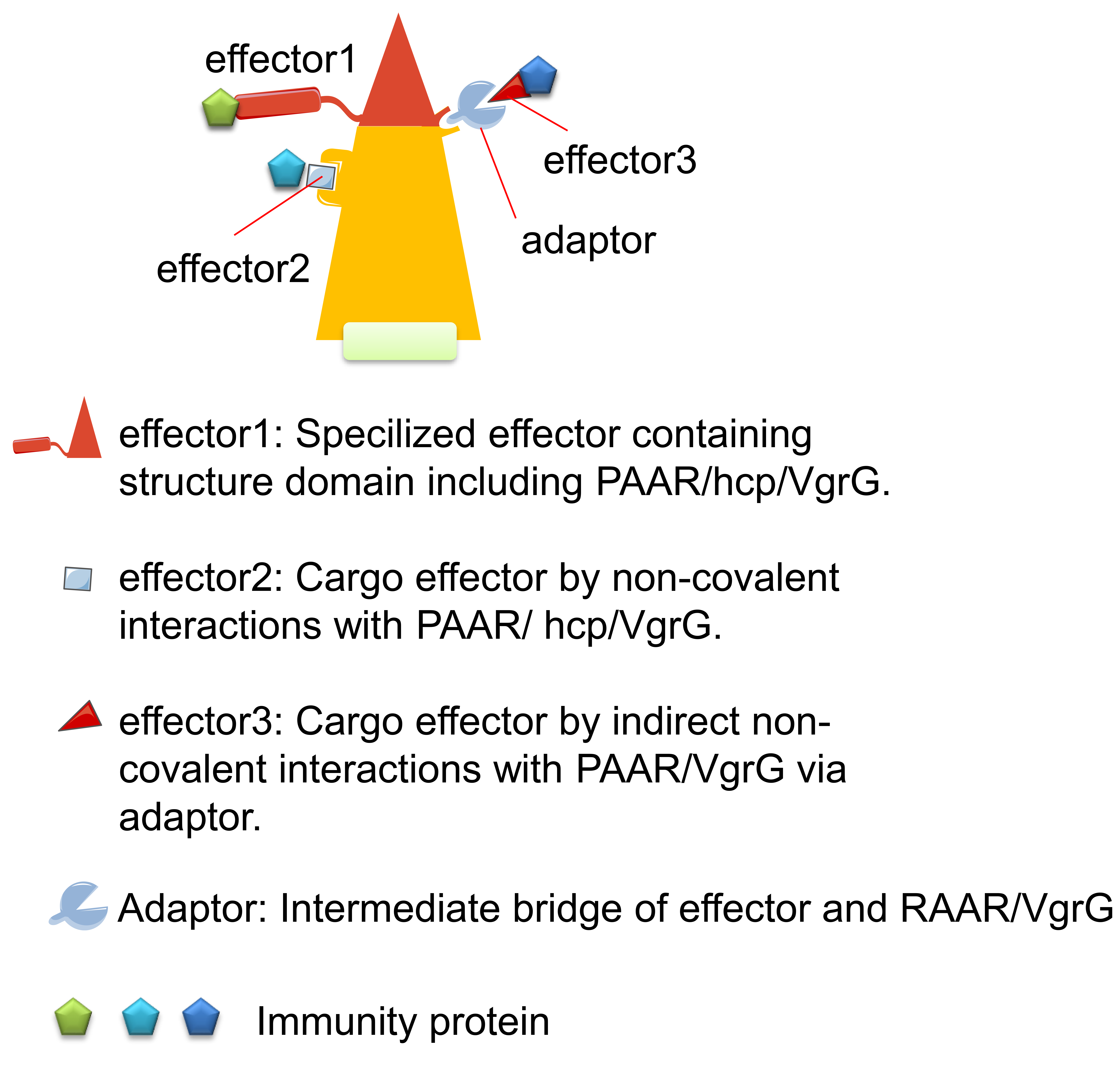
Overview of the T6SE vector assembly mode
T6SS data were exhibited by two entrances: Browse and Statistics. In the Statistics module, according to different filter conditions, T6SS data were exhibited in three portions: Classification, Taxonomy and Replicon. In Classification portion, data were further divided into T6SS classification, T6SS function type, T6SE, T6SR and T6SA for users' convenient browse. The whole T6SS data by experimental validated were exhibited in every organism via a summary thumbnail, users can view the information by clicking organism column, like Acinetobacter sp. ADP1 chromosome in T6SS list. The detailed information of T6SS and relationship between T6SS and T6SS-related proteins were showed by intuitive graph and diagram which users can get from T6SS ID column in T6SS list, like T6SS00061.
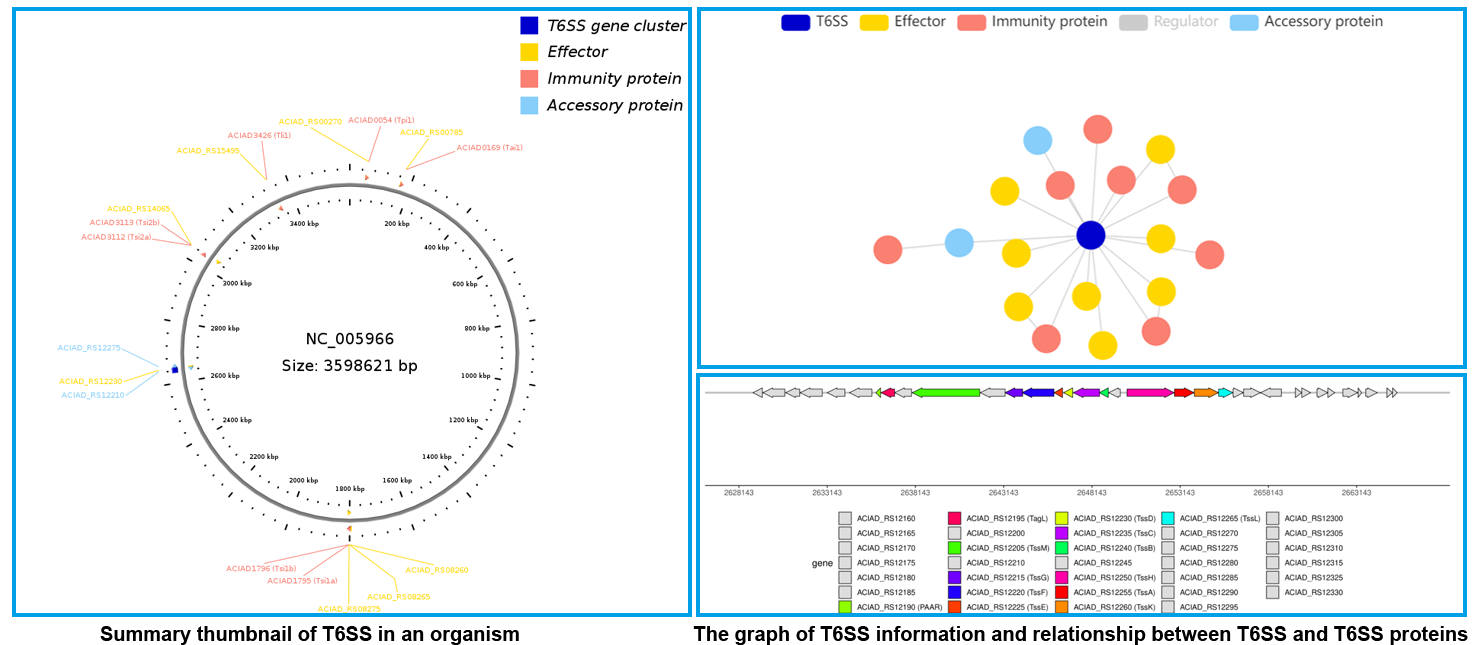
T6SS proteins were all exhibited detailed information and relationship with T6SS by intuitive graph and diagram through matching SecReT6 Protein ID or locus tag (Gene) in the sympathetic module, like Effector in Effector ID EFF01470, or locus tag (Gene) ACIAD_RS00785, which could be browsed in Effector list. Effectors were classified by function type from Tle to Tse, which users can enter into through T6SE portion in Classification module. T6SRs were classified by regulatory pathways in T6SR of Classification module. T6SA was constituted by adaptor, chaperone, and structure affiliated protein, also shown in T6SA of Classification module.
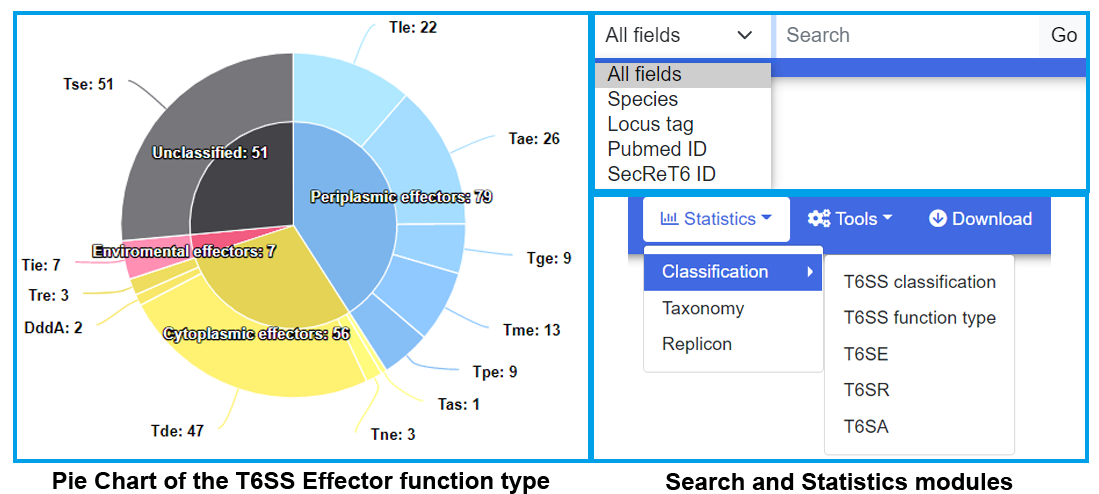
SecReT6 v3.0 serves a Search function in every webpage and data could be browsed through two entrances: Replicon or Taxonomy, to help users get information easily.
T6SS conserved components
| Component | Function | Cluster of Orthologous Groups |
| TssA | Unknown | COG3515 |
| TssB (IglA, VipA) | Tubular complex | COG3516 |
| TssC (IglB, VipB) | Tubular complex | COG3517 |
| TssD (Hcp) | Hexameric ring | COG3157 |
| TssE | Cytoplasmic protein | COG3518 |
| TssF | Unknown | COG3519 |
| TssG | Unknown | COG3520 |
| TssH (ClpV) | ATPase | COG0542 |
| TssI (VgrG) | Needle-like structure | COG3501 |
| TssJ | Transmembrane spanning structure | COG3521 |
| TssK | Cytoplasmic protein | COG3522 |
| TssL (DotU) | Transmembrane spanning structure | COG3455 |
| TssM (IcmF) | Transmembrane spanning structure | COG3523 |
More details at the Excel file.
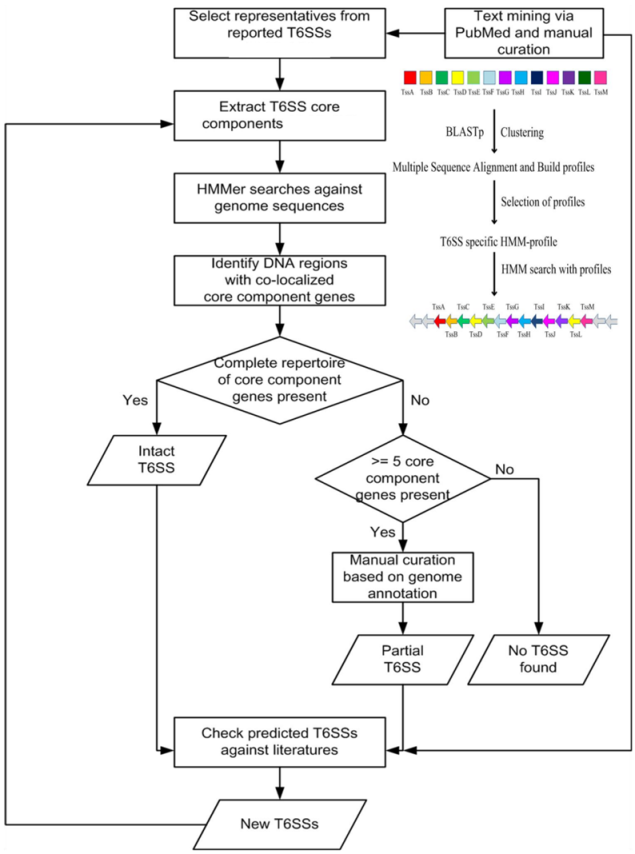
The prediction strategy used by SecReT6 to identify T6SS-encoding loci
References of T6SS assembly mode
[1] Bernal, P., et al. (2021). A novel stabilization mechanism for the type VI secretion system sheath. PNAS, 118(7), 1-9. https://doi.org/10.1073/pnas.2008500118[2] Wu, C.,et al. (2020). Effector loading onto the VgrG carrier activates type VI secretion system assembly. EMBO Reports, 21(1), 1-12. https://doi.org/10.15252/embr.201947961
[3] Wang, J., Brodmann, M., & Basler, M. (2019). Assembly and Subcellular Localization of Bacterial Type VI Secretion Systems. Annual Review of Microbiology, 73(1), 1-18. https://doi.org/10.1146/annurev-micro-020518-115420
[4] Pissaridou, P., et al. (2018). The Pseudomonas aeruginosa T6SS-VgrG1b spike is topped by a PAAR protein eliciting DNA damage to bacterial competitors. PNAS, 115 (49), 12519-12524. https://doi.org/10.1073/pnas.1814181115
[5] Burkinshaw, B. J., et al. (2018). A type VI secretion system effector delivery mechanism dependent on PAAR and a chaperone-co-chaperone complex. Nat Microbiol 3, 632-640. https://doi.org/10.1038/s41564-018-0144-4
[6] Yassine Cherrak Y, et al. (2018). Biogenesis and structure of a type VI secretion baseplate. Nat Microbiol 3, 1404-1416. https://doi.org/10.1038/s41564-018-0260-1
[7] Quentin, D., et al. (2018). Mechanism of loading and translocation of type VI secretion system effector Tse6. Nat Microbiol 3, 1142-1152. https://doi.org/10.1038/s41564-018-0238-z
[8] Unterweger, D., Kostiuk, B., & Pukatzki, S. (2017). Adaptor Proteins of Type VI Secretion System Effectors. Trends in Microbiology, 25(1), 8-10. https://doi.org/10.1016/j.tim.2016.10.003
[9] Nguyen, V. S., et al. (2017). Type VI secretion TssK baseplate protein exhibits structural similarity with phage receptor-binding proteins and evolved to bind the membrane complex. Nat Microbiol, 2, 1-9. https://doi.org/10.1038/nmicrobiol.2017.103
[10] Whitney, J. C., et al. (2015). An Interbacterial NAD(P)+ Glycohydrolase Toxin Requires Elongation Factor Tu for Delivery to Target Cells. Cell, 163(3), 607-619. https://doi.org/10.1016/j.cell.2015.09.027
[11] Durand, E., et al. (2015). Biogenesis and structure of a type VI secretion membrane core complex. Nature, 523(7562), 555-560. https://doi.org/10.1038/nature14667
[12] Mikhail M., et al. (2014). PAAR-repeat proteins sharpen and diversify the Type VI secretion system spike Mikhail. Nature, 500(7462), 350-353. https://doi.org/10.1038/nature12453