Detailed information of T6SS
Note: only experimentally validated proteins are displayed in the graph.
Summary
| T6SS ID | T6SS01168 |
| T6SS type | Type i1 |
| Strain | Vibrio cholerae C6706 |
| Replicon (RefSeq) | chromosome (NZ_CP046845) |
| Location | 296505..315674 |
| Function | anti-bacterial; anti-eukaryotic |
| Comment | T6SS is functional and T6SS-mediated killing causes an initially well-mixed population of mutually antagonistic bacteria to phase separate, forming and its VgrG-1 ACD is specifically necessary for the increased amplitude of intestinal contractions observed in fish monocolonized to alters the intestinal movements of larval zebrafish. |
| References | [1] Logan SL, Thomas J, Yan J, Baker RP, Shields DS, Xavier JB, Hammer BK, Parthasarathy R. et al (2018) The Vibrio cholerae type VI secretion system can modulate host intestinal mechanics to displace gut bacterial symbionts.. Proc Natl Acad Sci U S A. 115(16):E3779-E3787. [PudMed:29610339] |
T6SS components and genome coordinates
| Component | TssA | TssB | TssC | TssE | TssF | TssG | TssH | TssJ | TssK | TssL | TssM |
| Number | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
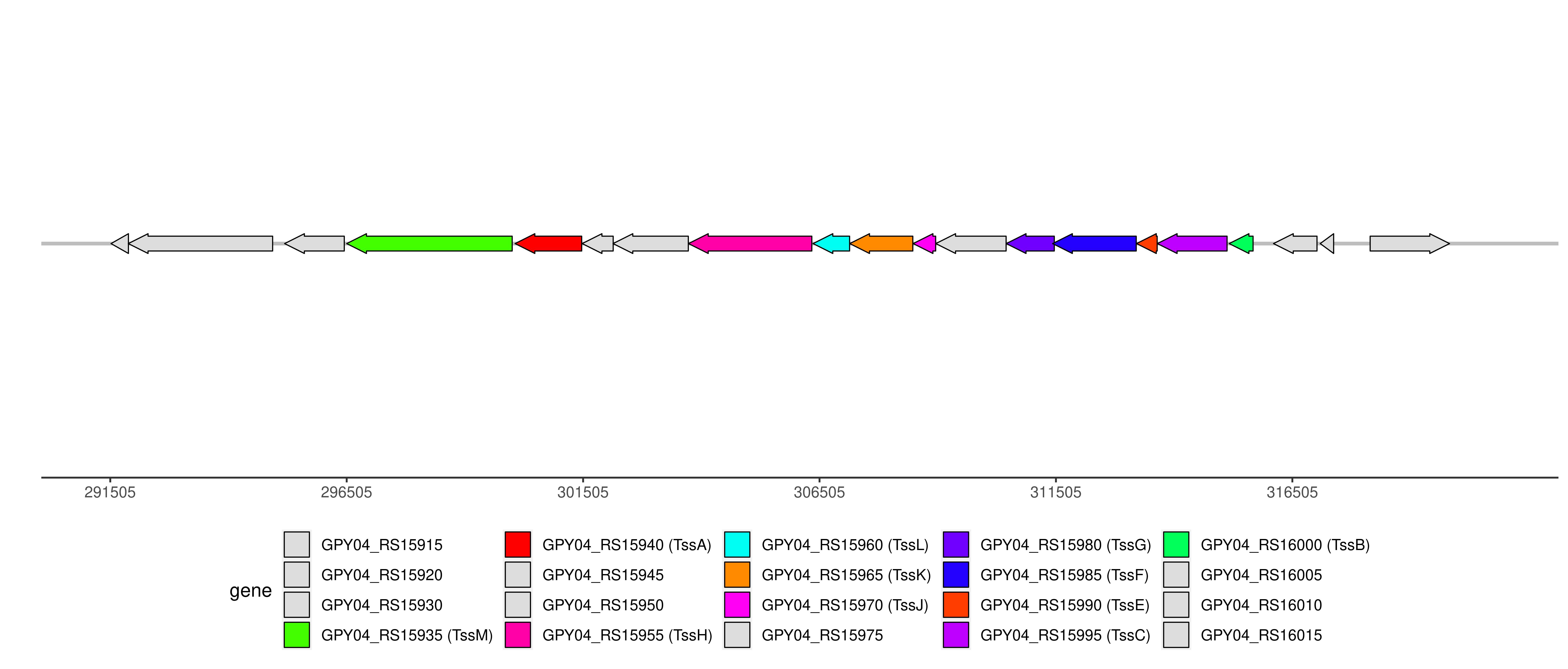
| Component ID | Component | Locus tag (Gene) | Coordinate | Strand | Size (bp) | NCBI ID | Product |
|---|---|---|---|---|---|---|---|
| GPY04_RS15915 | 291521..291889 | - | 369 | WP_001061295.1 | hypothetical protein | ||
| GPY04_RS15920 (tssI) | 291886..294939 | - | 3054 | WP_000113295.1 | type VI secretion system tip protein VgrG | ||
| GPY04_RS15930 | 295192..296454 | - | 1263 | WP_000073077.1 | ImpA family type VI secretion system protein | ||
| T6CP014862 | TssM | GPY04_RS15935 (tssM) | 296505..300005 | - | 3501 | WP_000225015.1 | type VI secretion system membrane subunit TssM |
| T6CP014863 | TssA | GPY04_RS15940 (vasJ) | 300066..301475 | - | 1410 | WP_000426093.1 | TssA family type VI secretion system protein VasJ |
| GPY04_RS15945 (tagO) | 301485..302141 | - | 657 | WP_001223388.1 | type VI secretion system-associated protein TagO | ||
| GPY04_RS15950 (vasH) | 302138..303730 | - | 1593 | WP_000084793.1 | sigma-54 dependent T6SS transcriptional regulator VasH | ||
| T6CP014864 | TssH | GPY04_RS15955 (tssH) | 303733..306342 | - | 2610 | WP_000619136.1 | type VI secretion system ATPase TssH |
| T6CP014865 | TssL | GPY04_RS15960 (icmH) | 306368..307141 | - | 774 | WP_000083409.1 | type IVB secretion system protein IcmH/DotU |
| T6CP014866 | TssK | GPY04_RS15965 (tssK) | 307144..308478 | - | 1335 | WP_000458007.1 | type VI secretion system baseplate subunit TssK |
| T6CP014867 | TssJ | GPY04_RS15970 (tssJ) | 308485..308961 | - | 477 | WP_001045311.1 | type VI secretion system lipoprotein TssJ |
| GPY04_RS15975 (tagH) | 308964..310451 | - | 1488 | WP_001089704.1 | type VI secretion system-associated FHA domain protein TagH | ||
| T6CP014868 | TssG | GPY04_RS15980 (tssG) | 310454..311470 | - | 1017 | WP_000510181.1 | type VI secretion system baseplate subunit TssG |
| T6CP014869 | TssF | GPY04_RS15985 (tssF) | 311434..313203 | - | 1770 | WP_000189792.1 | type VI secretion system baseplate subunit TssF |
| T6CP014870 | TssE | GPY04_RS15990 (tssE) | 313209..313646 | - | 438 | WP_000221000.1 | type VI secretion system baseplate subunit TssE |
| T6CP014871 | TssC | GPY04_RS15995 (tssC) | 313649..315124 | - | 1476 | WP_000108140.1 | type VI secretion system contractile sheath large subunit |
| T6CP014872 | TssB | GPY04_RS16000 (tssB) | 315168..315674 | - | 507 | WP_000031391.1 | type VI secretion system contractile sheath small subunit |
| GPY04_RS16005 | 316106..317026 | - | 921 | WP_001901866.1 | sel1 repeat family protein | ||
| GPY04_RS16010 | 317094..317378 | - | 285 | WP_000526403.1 | type VI secretion system PAAR protein | ||
| GPY04_RS16015 | 318150..319829 | + | 1680 | WP_001275247.1 | RluA family pseudouridine synthase |
Download FASTA format files
Note: in the 'Target' column, ↓ means the target is positively regulated by the regulator, while ↑ indicates negatively regulation.
| Effector ID | Locus tag (Gene) | Classification | Cognate immunity protein | Organism | Replicon (Accession) | Coordinates (Strand) | NCBI ID |
|---|---|---|---|---|---|---|---|
| EFF01818 |
GPY04_RS04830 (TleV1) | Tle | IMU00356 |
Vibrio cholerae C6706 | chromosome (NZ_CP046844) | 1067605..1069530 (+) | 446298981 |
| EFF01820 |
GPY04_RS16385 (VasX) | Tme | IMU00357 |
Vibrio cholerae C6706 | chromosome (NZ_CP046845) | 405867..409124 (-) | 445992499 |
| EFF01819 |
GPY04_RS04820 (VgrG-1) | - | - | Vibrio cholerae C6706 | chromosome (NZ_CP046844) | 1063272..1066763 (+) | 446134261 |
| EFF01821 |
GPY04_RS15920 | - | IMU00358 |
Vibrio cholerae C6706 | chromosome (NZ_CP046845) | 291886..294939 (-) | 446035440 |
| Immunity protein ID | Locus tag (Gene) | Cognate effector | Organism | Replicon (Accession) | Coordinates (Strand) | NCBI ID |
|---|---|---|---|---|---|---|
| IMU00356 |
GPY04_RS04835 (TsiV1) | EFF01818 |
Vibrio cholerae C6706 | chromosome (NZ_CP046844) | 1069527..1070270 (+) | 446694188 |
| IMU00357 |
GPY04_RS16380 (TsiV2) | EFF01820 |
Vibrio cholerae C6706 | chromosome (NZ_CP046845) | 405138..405866 (-) | 446826823 |
| IMU00358 |
GPY04_RS15915 (TsiV3) | EFF01821 |
Vibrio cholerae C6706 | chromosome (NZ_CP046845) | 291521..291889 (-) | 446984039 |
| Accessory protein ID | Locus tag (Gene) | Type | Organism | Replicon (Accession) | Coordinates (Strand) | NCBI ID | Related structure protein | Related effector |
|---|