Detailed information of T6SS
Note: only experimentally validated proteins are displayed in the graph.
Summary
| T6SS ID | T6SS00047 |
| T6SS type | Type i1 |
| Strain | Helicobacter hepaticus ATCC 51449 |
| Replicon (RefSeq) | chromosome (NC_004917) |
| Location | 227242..253067 |
| Function | anti-eukaryotic; effector translocation; virulence |
| Comment | Anti-inflammatory response in cultured intestinal epithelial cells and limitation of intestinal colonisation in mice (and perhaps therefore promotion of a tolerant, balanced relationship with host). |
| References | [1] Chow J et al (2010) A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe. 7(4):265-76. [PudMed:20413095] [2] Ge Z et al (2008) Helicobacter hepaticus HHGI1 is a pathogenicity island associated with typhlocolitis in B6.129-IL10 tm1Cgn mice. Microbes Infect. 10(7):726-33. [PudMed:18538610] [3] Bartonickova L et al (2013) Hcp and VgrG1 are secreted components of the Helicobacter hepaticus type VI secretion system and VgrG1 increases the bacterial colitogenic potential. Cell Microbiol. 15(6):992-1011. [PudMed:23278999] |
T6SS components and genome coordinates
| Component | FHA | TssB | TssC | TssD | TssF | TssG | TssI | TssJ | TssK | TssL | TssM |
| Number | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 |
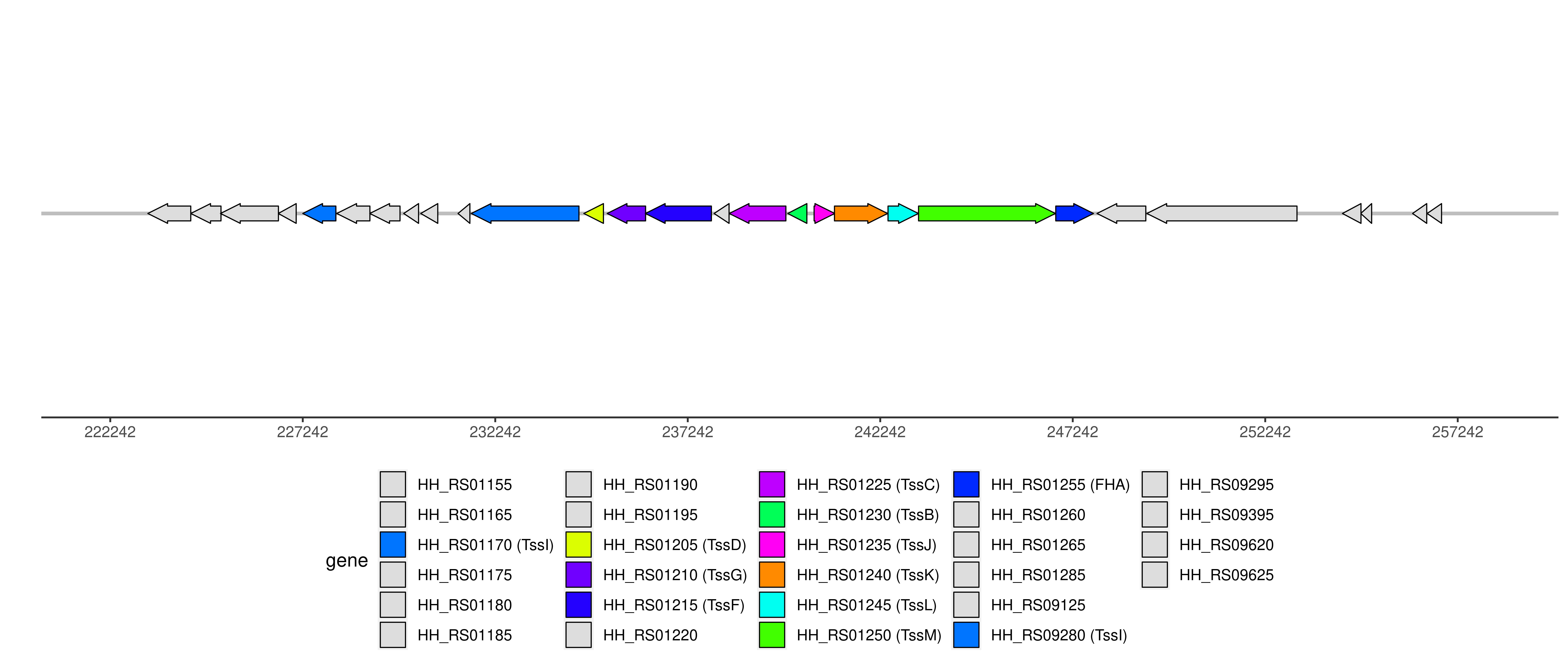
| Component ID | Component | Locus tag (Gene) | Coordinate | Strand | Size (bp) | NCBI ID | Product |
|---|---|---|---|---|---|---|---|
| HH_RS09620 | 223218..224333 | - | 1116 | WP_041308932.1 | hypothetical protein | ||
| HH_RS01155 | 224333..225118 | - | 786 | WP_011115078.1 | hypothetical protein | ||
| HH_RS09625 | 225105..226610 | - | 1506 | WP_011115079.1 | hypothetical protein | ||
| HH_RS01165 | 226607..227071 | - | 465 | WP_148141004.1 | hypothetical protein | ||
| T6CP009648 | TssI | HH_RS01170 | 227242..228102 | - | 861 | pseudogene | VgrG protein |
| HH_RS01175 | 228126..228986 | - | 861 | WP_011115082.1 | Dam family site-specific DNA-(adenine-N6)-methyltransferase | ||
| HH_RS01180 | 228983..229768 | - | 786 | WP_011115083.1 | EcoRV family type II restriction endonuclease | ||
| HH_RS01185 | 229864..230250 | - | 387 | WP_011115084.1 | hypothetical protein | ||
| HH_RS01190 | 230306..230749 | - | 444 | WP_148141034.1 | hypothetical protein | ||
| HH_RS01195 | 231275..231583 | - | 309 | pseudogene | hypothetical protein | ||
| T6CP000395 | TssI | HH_RS09280 | 231618..234416 | - | 2799 | WP_011115086.1 | hypothetical protein |
| T6CP008405 | TssD | HH_RS01205 | 234543..235052 | - | 510 | WP_011115087.1 | Hcp family type VI secretion system effector |
| T6CP000396 | TssG | HH_RS01210 (tssG) | 235149..236147 | - | 999 | WP_148141005.1 | type VI secretion system baseplate subunit TssG |
| T6CP000397 | TssF | HH_RS01215 (tssF) | 236141..237856 | - | 1716 | WP_011115089.1 | type VI secretion system baseplate subunit TssF |
| HH_RS01220 | 237926..238315 | - | 390 | WP_011115090.1 | hypothetical protein | ||
| T6CP000398 | TssC | HH_RS01225 (tssC) | 238319..239791 | - | 1473 | WP_011115091.1 | type VI secretion system contractile sheath large subunit |
| T6CP000399 | TssB | HH_RS01230 (tssB) | 239839..240336 | - | 498 | WP_011115092.1 | type VI secretion system contractile sheath small subunit |
| T6CP000400 | TssJ | HH_RS01235 (tssJ) | 240524..241060 | + | 537 | WP_011115093.1 | type VI secretion system lipoprotein TssJ |
| T6CP000401 | TssK | HH_RS01240 (tssK) | 241053..242432 | + | 1380 | WP_011115094.1 | type VI secretion system baseplate subunit TssK |
| T6CP000402 | TssL | HH_RS01245 | 242444..243232 | + | 789 | WP_011115095.1 | type IVB secretion system protein IcmH/DotU |
| T6CP008406 | TssM | HH_RS01250 (tssM) | 243237..246791 | + | 3555 | WP_011115096.1 | type VI secretion system membrane subunit TssM |
| T6CP011437 | FHA | HH_RS01255 | 246801..247781 | + | 981 | WP_011115097.1 | hypothetical protein |
| HH_RS01260 | 247872..249140 | - | 1269 | WP_011115098.1 | hypothetical protein | ||
| HH_RS01265 | 249168..253067 | - | 3900 | WP_011115099.1 | hypothetical protein | ||
| HH_RS09395 | 254247..254729 | - | 483 | pseudogene | hypothetical protein | ||
| HH_RS09125 | 254722..255006 | - | 285 | WP_050720625.1 | hypothetical protein | ||
| HH_RS01285 | 256069..256437 | - | 369 | WP_011115102.1 | hypothetical protein | ||
| HH_RS09295 | 256448..256822 | - | 375 | WP_083750358.1 | hypothetical protein |
Download FASTA format files
Note: in the 'Target' column, ↓ means the target is positively regulated by the regulator, while ↑ indicates negatively regulation.
| Regulator ID | Locus tag (Gene) | Regulator type | Regulatory pathway | Organism | Replicon (Accession) | Coordinates (Strand) | NCBI ID | Target (Regulation) |
|---|
| Effector ID | Locus tag (Gene) | Classification | Cognate immunity protein | Organism | Replicon (Accession) | Coordinates (Strand) | NCBI ID |
|---|---|---|---|---|---|---|---|
| EFF00064 |
HH_RS09280 | - | - | Helicobacter hepaticus ATCC 51449 | chromosome (NC_004917) | 231618..234416 (-) | 32265741 |
| EFF00065 |
HH_RS01205 | - | - | Helicobacter hepaticus ATCC 51449 | chromosome (NC_004917) | 234543..235052 (-) | 32265742 |
| Immunity protein ID | Locus tag (Gene) | Cognate effector | Organism | Replicon (Accession) | Coordinates (Strand) | NCBI ID |
|---|
| Accessory protein ID | Locus tag (Gene) | Type | Organism | Replicon (Accession) | Coordinates (Strand) | NCBI ID | Related structure protein | Related effector |
|---|