Detailed information of T6SS
Note: only experimentally validated proteins are displayed in the graph.
Summary
| T6SS ID | T6SS00041 |
| T6SS type | Type i1 |
| Strain | Pseudomonas syringae pv. tomato str. DC3000 |
| Replicon (RefSeq) | chromosome (NC_004578) |
| Location | 2796816..2829435 |
| Function | - |
| Comment | - |
| References | [1] Haapalainen M et al (2012) Hcp2, a Secreted Protein of the Phytopathogen Pseudomonas syringae pv. Tomato DC3000, Is Required for Fitness for Competition against Bacteria and Yeasts. J Bacteriol. 194(18):4810-22. [PudMed:22753062] [2] Sarris PF et al (2010) In silico analysis reveals multiple putative type VI secretion systems and effector proteins in Pseudomonas syringae pathovars. Mol Plant Pathol. 11(6):795-804. [PudMed:21091602] [3] Boyer F et al (2009) Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources. BMC Genomics. 0.488888889. [PudMed:19284603] [4] Barret M et al (2011) Genomic analysis of the type VI secretion systems in Pseudomonas spp.: novel clusters and putative effectors uncovered. Microbiology. 157(Pt 6):1726-39. [PudMed:21474537] |
T6SS components and genome coordinates
| Component | Sfa2 | TssB | TssC | TssD | TssE | TssF | TssG | TssH | TssI | TssJ | TssK | TssL | TssM |
| Number | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
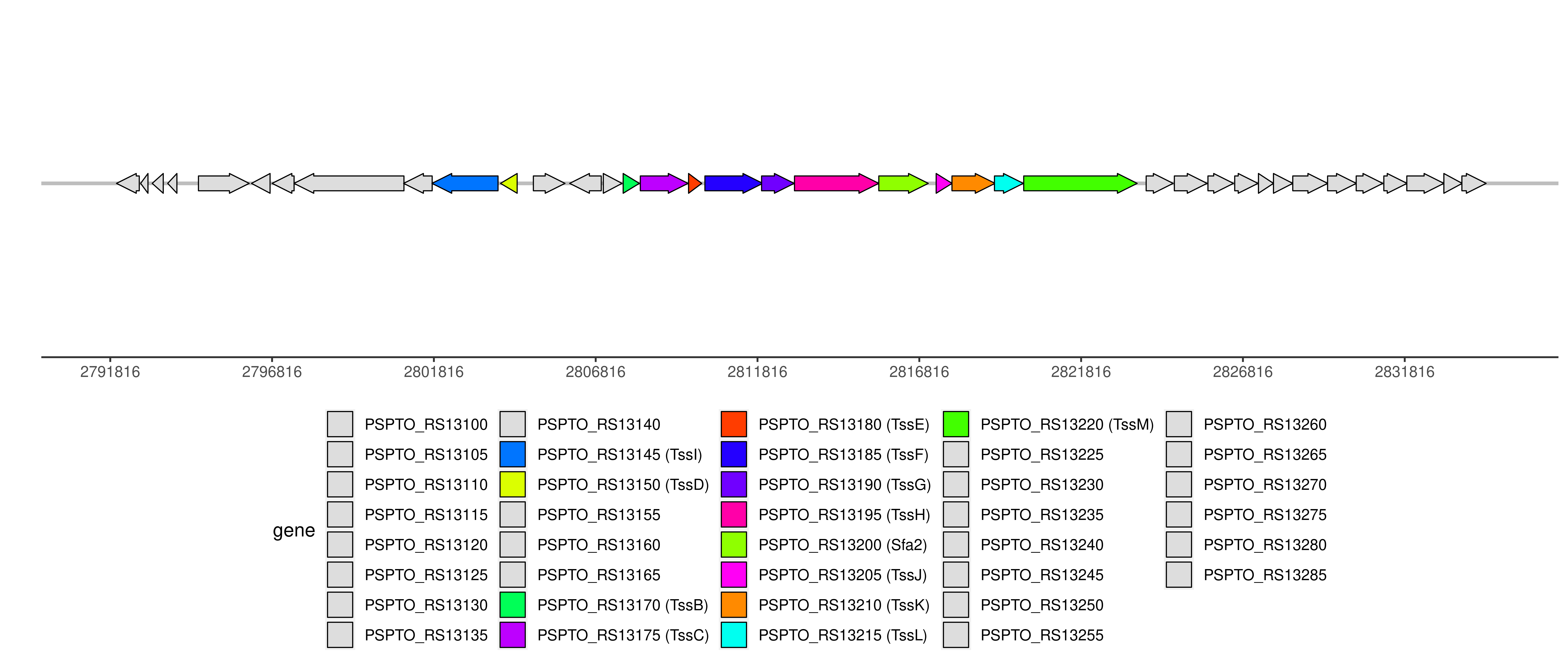
| Component ID | Component | Locus tag (Gene) | Coordinate | Strand | Size (bp) | NCBI ID | Product |
|---|---|---|---|---|---|---|---|
| PSPTO_RS13100 | 2792008..2792712 | - | 705 | WP_044393213.1 | DUF1275 domain-containing protein | ||
| PSPTO_RS13105 | 2792754..2792981 | - | 228 | WP_003383427.1 | hypothetical protein | ||
| PSPTO_RS13110 | 2793112..2793453 | - | 342 | WP_007246648.1 | FKBP-type peptidyl-prolyl cis-trans isomerase | ||
| PSPTO_RS13115 | 2793591..2793875 | - | 285 | WP_011104063.1 | hypothetical protein | ||
| PSPTO_RS13120 | 2794541..2796109 | + | 1569 | WP_011104064.1 | TerC family protein | ||
| PSPTO_RS13125 | 2796179..2796754 | - | 576 | WP_011104065.1 | GNAT family N-acetyltransferase | ||
| PSPTO_RS13130 | 2796816..2797496 | - | 681 | WP_011104066.1 | hypothetical protein | ||
| PSPTO_RS13135 | 2797493..2800891 | - | 3399 | WP_164923175.1 | hypothetical protein | ||
| PSPTO_RS13140 | 2800881..2801763 | - | 883 | pseudogene | DUF4123 domain-containing protein | ||
| T6CP000334 | TssI | PSPTO_RS13145 | 2801756..2803798 | - | 2043 | WP_011104070.1 | type VI secretion system tip protein VgrG |
| T6CP008396 | TssD | PSPTO_RS13150 | 2803874..2804389 | - | 516 | WP_007246961.1 | Hcp family type VI secretion system effector |
| PSPTO_RS13155 (tssA) | 2804890..2805867 | + | 978 | pseudogene | type VI secretion system protein TssA | ||
| PSPTO_RS13160 | 2806011..2806988 | - | 978 | WP_007247571.1 | IS5-like element ISPssy family transposase | ||
| PSPTO_RS13165 | 2807051..2807629 | + | 579 | pseudogene | type VI secretion system domain-containing protein | ||
| T6CP000335 | TssB | PSPTO_RS13170 (tssB) | 2807665..2808165 | + | 501 | WP_011104071.1 | type VI secretion system contractile sheath small subunit |
| T6CP000336 | TssC | PSPTO_RS13175 (tssC) | 2808195..2809670 | + | 1476 | WP_011104072.1 | type VI secretion system contractile sheath large subunit |
| T6CP008397 | TssE | PSPTO_RS13180 (tssE) | 2809686..2810093 | + | 408 | WP_011104073.1 | type VI secretion system baseplate subunit TssE |
| T6CP000337 | TssF | PSPTO_RS13185 (tssF) | 2810191..2811978 | + | 1788 | WP_011104074.1 | type VI secretion system baseplate subunit TssF |
| T6CP000338 | TssG | PSPTO_RS13190 (tssG) | 2811942..2812949 | + | 1008 | WP_011104075.1 | type VI secretion system baseplate subunit TssG |
| T6CP000339 | TssH | PSPTO_RS13195 (tssH) | 2812959..2815562 | + | 2604 | WP_011104076.1 | type VI secretion system ATPase TssH |
| T6CP012435 | Sfa2 | PSPTO_RS13200 | 2815564..2817093 | + | 1530 | WP_011104077.1 | sigma-54-dependent Fis family transcriptional regulator |
| T6CP000340 | TssJ | PSPTO_RS13205 (tssJ) | 2817337..2817828 | + | 492 | WP_007246952.1 | type VI secretion system lipoprotein TssJ |
| T6CP000341 | TssK | PSPTO_RS13210 (tssK) | 2817825..2819156 | + | 1332 | WP_010213774.1 | type VI secretion system baseplate subunit TssK |
| T6CP000342 | TssL | PSPTO_RS13215 | 2819138..2820028 | + | 891 | WP_046463302.1 | type IVB secretion system protein IcmH/DotU |
| T6CP000343 | TssM | PSPTO_RS13220 (tssM) | 2820042..2823548 | + | 3507 | WP_011104079.1 | type VI secretion system membrane subunit TssM |
| PSPTO_RS13225 (phnC) | 2823826..2824659 | + | 834 | WP_011104080.1 | phosphonate ABC transporter ATP-binding protein | ||
| PSPTO_RS13230 (phnD) | 2824697..2825707 | + | 1011 | WP_007246947.1 | phosphonate ABC transporter substrate-binding protein | ||
| PSPTO_RS13235 (phnE) | 2825735..2826523 | + | 789 | WP_007246946.1 | phosphonate ABC transporter, permease protein PhnE | ||
| PSPTO_RS13240 (phnF) | 2826562..2827281 | + | 720 | WP_011104081.1 | phosphonate metabolism transcriptional regulator PhnF | ||
| PSPTO_RS13245 (phnG) | 2827296..2827760 | + | 465 | WP_007246944.1 | phosphonate C-P lyase system protein PhnG | ||
| PSPTO_RS13250 (phnH) | 2827760..2828356 | + | 597 | WP_011104082.1 | phosphonate C-P lyase system protein PhnH | ||
| PSPTO_RS13255 | 2828356..2829435 | + | 1080 | WP_011104083.1 | carbon-phosphorus lyase complex subunit PhnI | ||
| PSPTO_RS13260 | 2829432..2830319 | + | 888 | WP_011104084.1 | alpha-D-ribose 1-methylphosphonate 5-phosphate C-P-lyase PhnJ | ||
| PSPTO_RS13265 (phnK) | 2830316..2831137 | + | 822 | WP_011104085.1 | phosphonate C-P lyase system protein PhnK | ||
| PSPTO_RS13270 (phnL) | 2831170..2831886 | + | 717 | WP_011104086.1 | phosphonate C-P lyase system protein PhnL | ||
| PSPTO_RS13275 | 2831880..2833025 | + | 1146 | WP_011104087.1 | alpha-D-ribose 1-methylphosphonate 5-triphosphate diphosphatase | ||
| PSPTO_RS13280 (phnN) | 2833025..2833591 | + | 567 | WP_011104088.1 | phosphonate metabolism protein/1,5-bisphosphokinase (PRPP-forming) PhnN | ||
| PSPTO_RS13285 (phnP) | 2833582..2834331 | + | 750 | WP_011104089.1 | phosphonate metabolism protein PhnP |
Download FASTA format files
Note: in the 'Target' column, ↓ means the target is positively regulated by the regulator, while ↑ indicates negatively regulation.
| Regulator ID | Locus tag (Gene) | Regulator type | Regulatory pathway | Organism | Replicon (Accession) | Coordinates (Strand) | NCBI ID | Target (Regulation) |
|---|---|---|---|---|---|---|---|---|
| REG0096 |
PSPTO_RS08845 (relA) | protein | (p)ppGpp regulatory pathway | Pseudomonas syringae pv. tomato str. DC3000 | chromosome (NC_004578) | 1863879..1866122 (+) | 494457621 | T6SS transcripts (↓) |
| REG0097 |
PSPTO_RS00410 (spoT) | protein | (p)ppGpp regulatory pathway | Pseudomonas syringae pv. tomato str. DC3000 | chromosome (NC_004578) | 94805..96910 (-) | 492171753 | T6SS transcripts (↓) |
| REG0098 |
PSPTO_RS04545 (fpRel) | protein | (p)ppGpp regulatory pathway | Pseudomonas syringae pv. tomato str. DC3000 | chromosome (NC_004578) | 927082..927900 (+) | 499415849 | T6SS transcripts (↓) |
| REG0099 |
PSPTO_RS28060 (Sfa) | protein | Global regulator | Pseudomonas syringae pv. tomato str. DC3000 | chromosome (NC_004578) | 6165047..6166564 (-) | 499418021 | T6SS transcripts (↓) |
| Effector ID | Locus tag (Gene) | Classification | Cognate immunity protein | Organism | Replicon (Accession) | Coordinates (Strand) | NCBI ID |
|---|
| Immunity protein ID | Locus tag (Gene) | Cognate effector | Organism | Replicon (Accession) | Coordinates (Strand) | NCBI ID |
|---|
| Accessory protein ID | Locus tag (Gene) | Type | Organism | Replicon (Accession) | Coordinates (Strand) | NCBI ID | Related structure protein | Related effector |
|---|