
| 225 | |
| Bartonella tribocorum CIP 105476 | |
| chromosome [Browse all T4SS(s) in this replicon] | |
| NC_010161 | |
| 1800215..1812931 | |
| VirB | |
| effector translocation | |
| Type IVA; Type P | |
T4SS components |
| Component | VirB1 | VirB2 | VirB3 | VirB4 | VirB5 | VirB6 | VirB7 | VirB8 | VirB9 |
| Number | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Component | VirB10 | VirB11 | VirD4 | ||||||
| Number | 1 | 1 | 1 |
The information of T4SS components from NC_010161 | ||||||
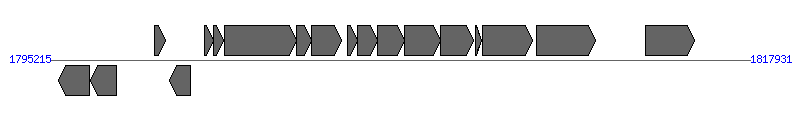 | ||||||
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | Btr_1685 (ribF) | 1795506..1796489 [-], 984 | 163868770 | bifunctional riboflavin kinase/FMN adenylyltransferase | ||
| 2 | Btr_1686 | 1796522..1797367 [-], 846 | 163868771 | HAD family sugar phosphatase | ||
| 3 | Btr_1687 (dicA) | 1798604..1798963 [+], 360 | 163868772 | transcriptional regulator | ||
| 4 | Btr_1688 | 1799101..1799772 [-], 672 | 163868773 | hypothetical protein | ||
| 5 | Btr_1689 (virB2) | 1800215..1800535 [+], 321 | 163868774 | VirB2 protein | VirB2 | |
| 6 | Btr_1690 (virB3) | 1800536..1800844 [+], 309 | 163868775 | VirB3 protein | VirB3 | |
| 7 | Btr_1691 (virB4) | 1800867..1803221 [+], 2355 | 163868776 | VirB4 protein | VirB4 | |
| 8 | Btr_1692 (virB5) | 1803218..1803703 [+], 486 | 163868777 | VirB5 protein | VirB5 | |
| 9 | Btr_1693 (virB6) | 1803690..1804667 [+], 978 | 163868778 | VirB6 protein | VirB6 | |
| 10 | Btr_1694 (virB7) | 1804879..1805193 [+], 315 | 163868779 | VirB7 protein | VirB7 | |
| 11 | Btr_1695 (virB8) | 1805183..1805851 [+], 669 | 163868780 | VirB8 protein | VirB8 | |
| 12 | Btr_1696 (virB9) | 1805848..1806714 [+], 867 | 163868781 | VirB9 protein precursor | VirB9 | |
| 13 | Btr_1697 (virB10) | 1806707..1807879 [+], 1173 | 163868782 | VirB10 protein | VirB10 | |
| 14 | Btr_1698 (virB11) | 1807876..1808946 [+], 1071 | 163868783 | VirB11 protein | VirB11 | |
| 15 | Btr_1699 | 1809009..1809224 [+], 216 | 163868784 | hypothetical protein | ||
| 16 | Btr_1700 (bepA) | 1809235..1810869 [+], 1635 | 163868785 | hypothetical protein | ||
| 17 | Btr_1701 (virD4) | 1811015..1812931 [+], 1917 | 163868786 | conjugal transfer coupling protein TraG | VirD4 | |
| 18 | Btr_1704 (bepC) | 1814544..1816142 [+], 1599 | 163868787 | hypothetical protein | ||
Download FASTA format files |
| Proteins Genes |
Effectors |
| BepA, BepC, BepD, BepH, BepE, BepF, BepI |
The information of protein effectors | ||||||
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | * | |
| 1 | Btr_1700 (bepA) | 1809235..1810869 [+], 1635 | 163868785 | hypothetical protein | BepA | |
| 2 | Btr_1704 (bepC) | 1814544..1816142 [+], 1599 | 163868787 | hypothetical protein | BepC | |
| 3 | Btr_1705 (bepD) | 1816452..1818026 [+], 1575 | 163868788 | hypothetical protein | BepD | |
| 4 | Btr_1706 (bepH) | 1818370..1819188 [+], 819 | 163868789 | BepH protein | BepH | |
| 5 | Btr_1707 (bepE) | 1819541..1820833 [+], 1293 | 163868790 | hypothetical protein | BepE | |
| 6 | Btr_1709 (bepF) | 1821138..1823105 [+], 1968 | 163868791 | hypothetical protein | BepF | |
| 7 | Btr_1710 (bepI) | 1823441..1825813 [+], 2373 | 163868792 | BepI protein | BepI | |
Download FASTA format files |
| Proteins Genes |
| # | Name | Image | Resource | Detail | Reference |
| 1 | VirB8 | 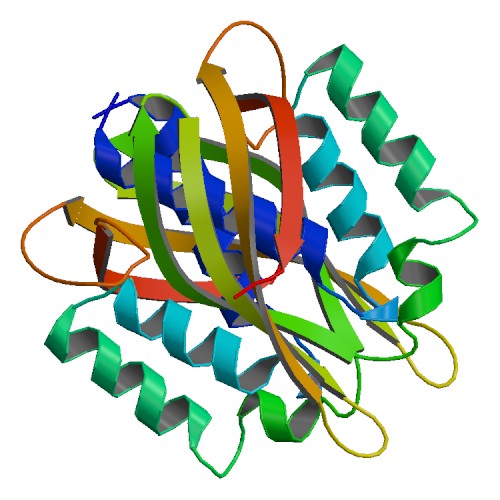 | PDB (4MEI) | Crystal structure of a VirB8 Type IV secretion system machinery soluble domain from Bartonella tribocorum. | (1) PubMed: 26646013 |
|
(1) Gillespie JJ et al. (2015). Structural Insight into How Bacteria Prevent Interference between Multiple Divergent Type IV Secretion Systems. MBio. 6(6):e01867-15. [PudMed:26646013] |
| (1) Schulein R; Dehio C (2002). The VirB/VirD4 type IV secretion system of Bartonella is essential for establishing intraerythrocytic infection. Mol Microbiol. 46(4):1053-67. [PudMed:12421311] |