Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100181 |
| Name | oriT_ColE1 |
| Organism | Escherichia coli |
| Sequence Completeness | core |
| NCBI accession of oriT (coordinates [strand]) | NC_001371 (1462..1521 [-], 60 nt) |
| oriT length | 60 nt |
| IRs (inverted repeats) | 3..12, 16..25 (GTGTCGGGGC..GCCCTGACCC) |
| Location of nic site | 56..57 |
| Conserved sequence flanking the nic site |
CTGG|CTTA |
| Note | The R13 amino residue of auxiliary protein MbeC_ColE1 is essential to the DNA-binding activity [PMID:19114496]. |
oriT sequence
Download Length: 60 nt
GGGTGTCGGGGCGCAGCCCTGACCCAGTCACGTAGCGATAGCGGAGTGTATACTGGCTTA
Visualization of oriT structure (The oriT was characterized experimentally)
oriT secondary structure
Predicted by RNAfold.
Download structure file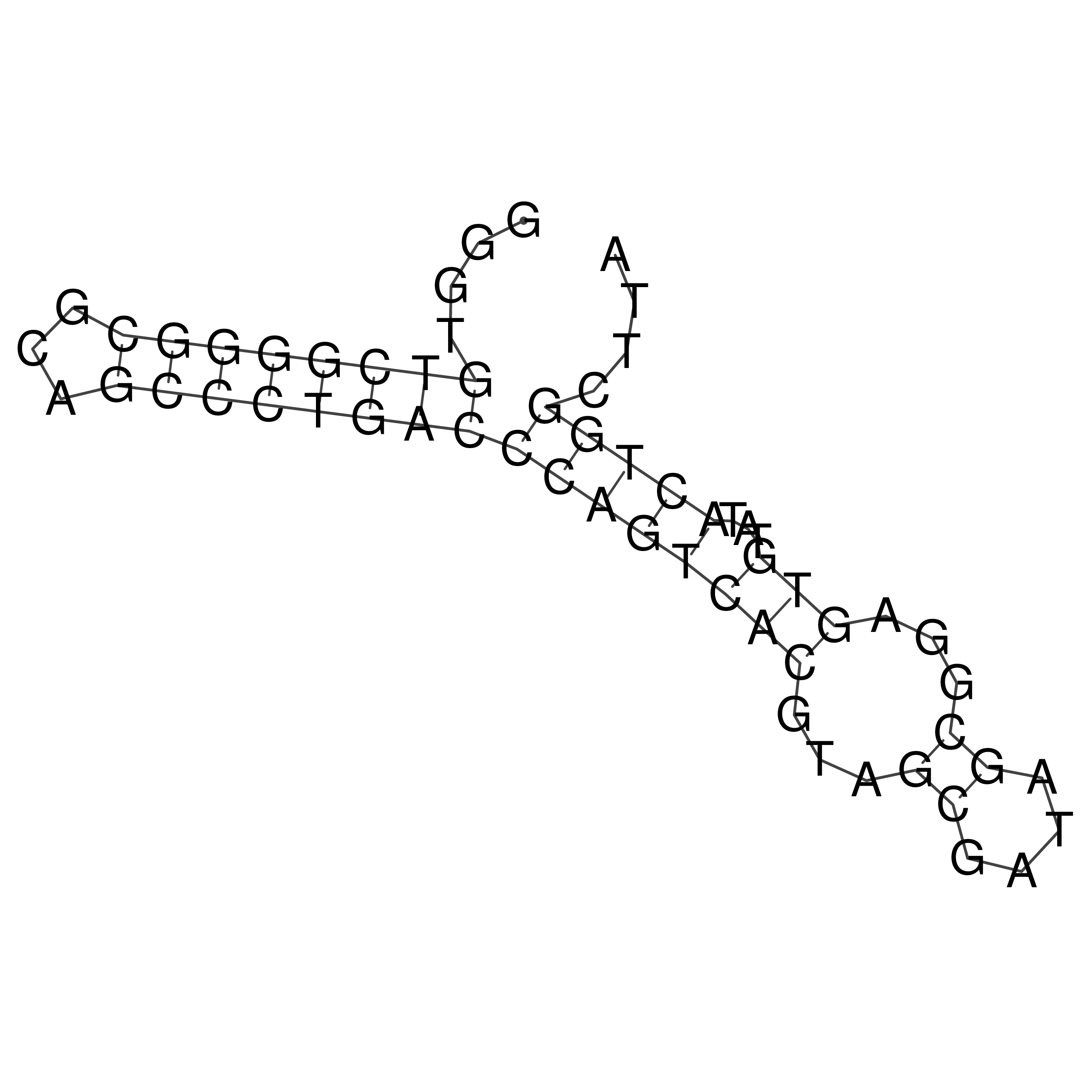
Reference
[1] Rogge ML et al. (2013) Comparison of Vietnamese and US isolates of Edwardsiella ictaluri. Dis Aquat Organ. 106(1):17-29. [PMID:24062549]
[2] Varsaki A et al. (2009) Analysis of ColE1 MbeC unveils an extended ribbon-helix-helix family of nicking accessory proteins. J Bacteriol. 191(5):1446-55. [PMID:19114496]
[3] Francia MV et al. (2004) A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol Rev. 28(1):79-100. [PMID:14975531]
Relaxosome
This oriT is a component of a relaxosome.
| Relaxosome name | RelaxosomeColE1 |
| oriT | oriT_ColE1 |
| Relaxase | MbeA_ColE1 |
| Auxiliary protein | MbeB_ColE1 |
Reference
[1] Rogge ML et al. (2013) Comparison of Vietnamese and US isolates of Edwardsiella ictaluri. Dis Aquat Organ. 106(1):17-29. [PMID:24062549]
[2] Varsaki A et al. (2009) Analysis of ColE1 MbeC unveils an extended ribbon-helix-helix family of nicking accessory proteins. J Bacteriol. 191(5):1446-55. [PMID:19114496]
[3] Francia MV et al. (2004) A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol Rev. 28(1):79-100. [PMID:14975531]
Relaxase
| ID | 174 | GenBank | CAA33883 |
| Name | MbeA_ColE1 |
UniProt ID | P13658 |
| Length | 517 a.a. | PDB ID | |
| Note | relaxase | ||
Relaxase protein sequence
Download Length: 517 a.a. Molecular weight: 57808.29 Da Isoelectric Point: 6.0811
MIVKFHARGKGGGSGPVDYLLGRERNREGATVLQGNPEEVRELIDATPFAKKYTSGVLSFAEKELPPGGR
EKVMASFERVLMPGLEKNQYSILWVEHQDKGRLELNFVIPNMELQTGKRLQPYYDRADRPRIDAWQTLVN
HHYGLHDPNAPENRRTLTLPDNLPETKQALAEGVTRGIDALYHAGEIKGRQDVIQALTEAGLEVVRVTRT
SISIADPNGGKNIRLKGAFYEQSFADGRGVREKAERESRIYRENAEQRVQEARRICKRGCDIKRDENQRR
YSPVHSLDRGIAGKTPGRGERGDDAAQEGRVKAGREYGHDVTGDSLSPVYREWRDALVSWREDTGEPGRN
QEAGRDIAETEREDMGRGVCAGREQEIPCPSVREISGGDSLSGERVGTSEGVTQSDRAGNTFAERLRAAA
TGLYAAAERMGERLRGIAEDVFAYATGQRDAERAGHAVESAGAALERADRTLEPVIQRELEIREERLIQE
REHVLSLERERQPEIQERTLDGPSLGW
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | P13658 |
Reference
[1] Varsaki A et al. (2009) Analysis of ColE1 MbeC unveils an extended ribbon-helix-helix family of nicking accessory proteins. J Bacteriol. 191(5):1446-55. [PMID:19114496]
[2] Francia MV et al. (2004) A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol Rev. 28(1):79-100. [PMID:14975531]
Auxiliary protein
| ID | 250 | GenBank | CAA33884 |
| Name | MbeB_ColE1 |
UniProt ID | P13659 |
| Length | 172 a.a. | PDB ID | _ |
| Note | _ | ||
Auxiliary protein sequence
Download Length: 172 a.a. Molecular weight: 19547.43 Da Isoelectric Point: 7.3220
MSNLLQTGAEFEKKLKERAESTEKMLNNEFRRLGESVSEAVTSNETKIRDAIALFTASTEESLEKHREGV
KEAMMQHRRDVLKLAGNTGMMLLGIVFLLFTASGGTLWYLGGRIQANLEEIRKQEETLQKLNAKTWGVEF
VQDGNRKFLVLPYGKSAEVIPFQGKEWVHLKE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | P13659 |
Reference
[1] Varsaki A et al. (2009) Analysis of ColE1 MbeC unveils an extended ribbon-helix-helix family of nicking accessory proteins. J Bacteriol. 191(5):1446-55. [PMID:19114496]
[2] Francia MV et al. (2004) A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol Rev. 28(1):79-100. [PMID:14975531]
| ID | 251 | GenBank | CAA33882 |
| Name | MbeC_ColE1 |
UniProt ID | P13657 |
| Length | 107 a.a. | PDB ID | _ |
| Note | The R13 amino residue of auxiliary protein MbeC_ColE1 is essential to the DNA-binding activity [PMID:19114496]. | ||
Auxiliary protein sequence
Download Length: 107 a.a. Molecular weight: 11855.71 Da Isoelectric Point: 10.8973
MLTIRVTDDEHARLLERCEGKQLAVWMRRVCLGEPVARSGKLPTLAPPLLRQLAAIGNNLNQTARKVNSG
QWSSGDRVQVVAALMAIGDELRRLRLAVREQGARDDS
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | P13657 |
Reference
[1] Varsaki A et al. (2009) Analysis of ColE1 MbeC unveils an extended ribbon-helix-helix family of nicking accessory proteins. J Bacteriol. 191(5):1446-55. [PMID:19114496]
[2] Francia MV et al. (2004) A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol Rev. 28(1):79-100. [PMID:14975531]
Host bacterium
| ID | 174 | GenBank | NC_001371 |
| Plasmid name | ColE1 | Incompatibility group | ColRNAI |
| Plasmid size | 6646 bp | Coordinate of oriT [Strand] | 1462..1521 [-] |
| Host baterium | Escherichia coli |
Cargo genes
| Drug resistance gene | _ |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |