Help
Introduction
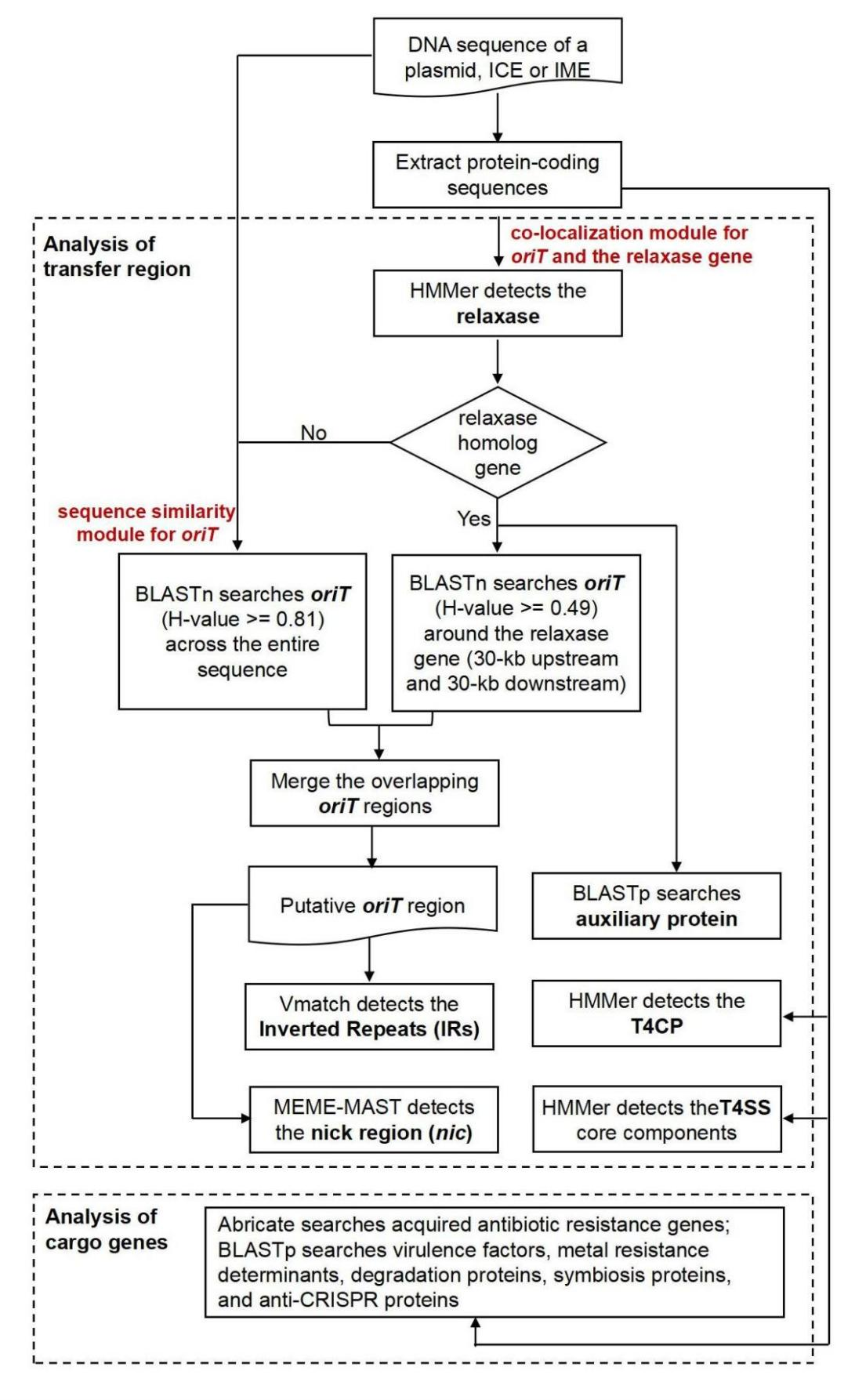
Bacterial mobile genetic elements (MGEs), play a critical role in disseminating pathogenesis and antimicrobial-resistance determinants. These elements, such as conjugative plasmids, Integrative and Conjugative Elements (ICEs), and Integrative and Mobilizable Elements (IMEs), typically consist of four modules in their conjugative regions: the origin-of-transfer (oriT) region, relaxase gene, type IV coupling protein (T4CP) gene, a gene cluster coding for bacterial type IV secretion system (T4SS), as well as the cargo genes in the MGEs. Focusing on the conjugative modules, oriTDB currently contains comprehensive information on the oriT regions, relaxases, auxiliary proteins, T4CP, T4SS and their associations in MGEs.
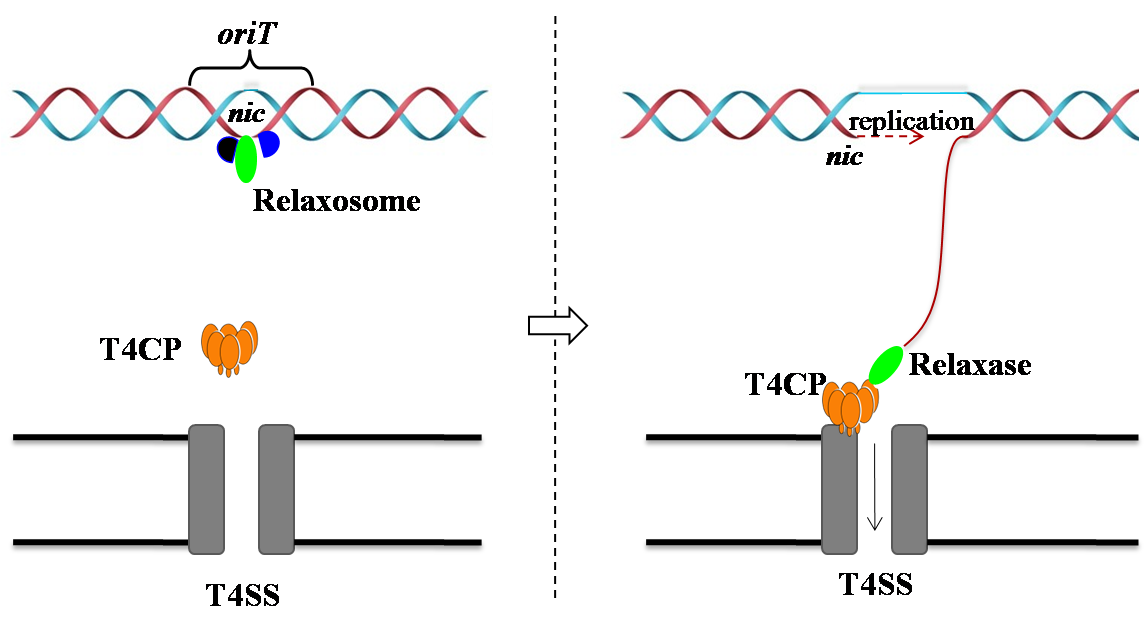
Key features
Key functions
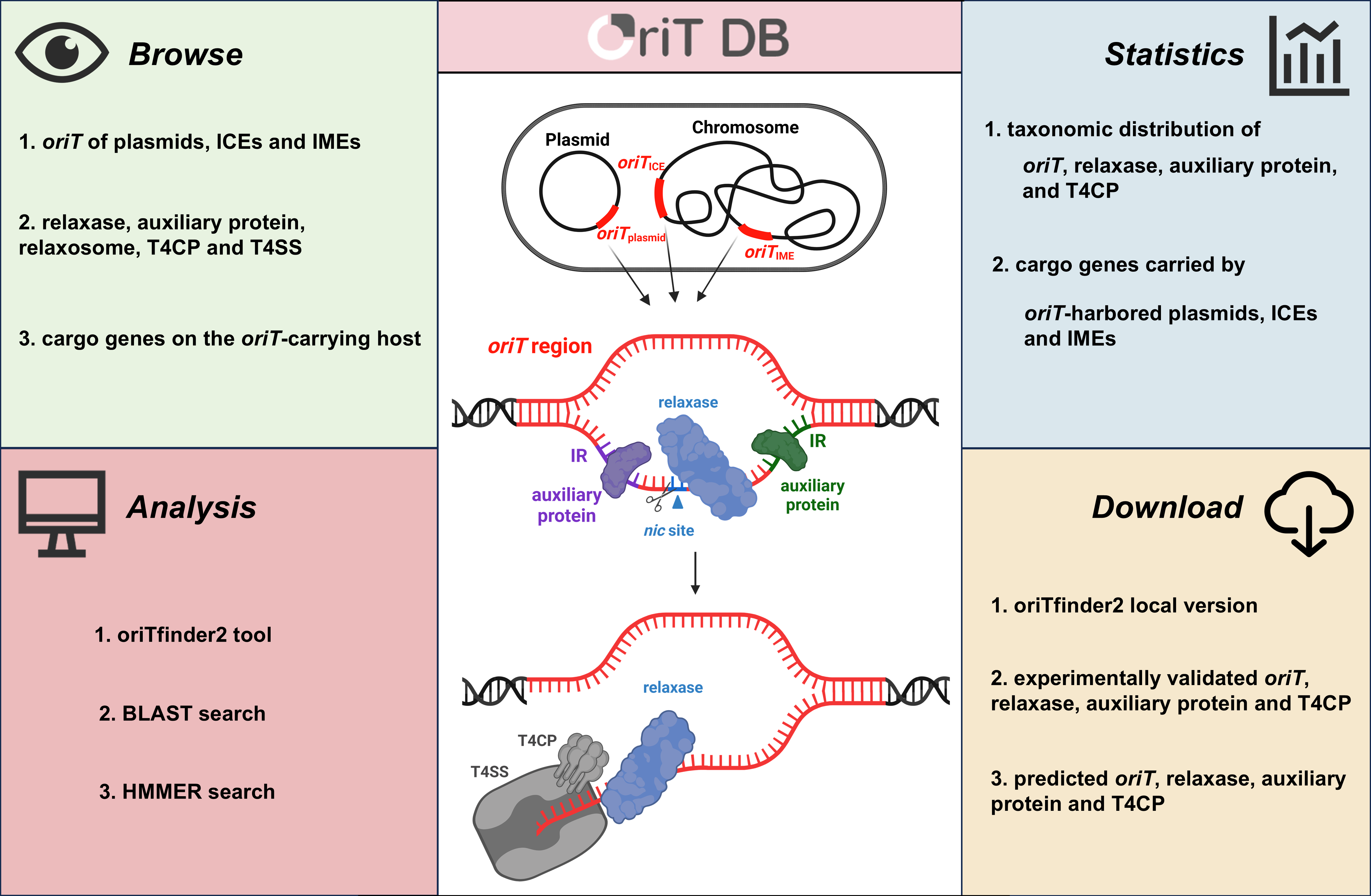
Data collection
Compared to the oriTDB 1.0 released in 2018, we updated our datasets by text mining and in silico prediction:
| Number of elements derived from experimental data | Number of elements derived from predicted data | Total | ||
| oriTDB (released in 2018) | ||||
| oriT | Plasmid | 42 | 954 | 996 |
| ICE | 6 | 67 | 73 | |
| IME | 4 | 2 | 6 | |
| Relaxase | 27 | 956 | 983 | |
| Auxiliary protein | 29 | 73 | 102 | |
| T4CP | 12 | 452 | 464 | |
| oriTDB (updated in 2024) | ||||
| oriT | Plasmid | 91 | 22,390 | 22,481 |
| ICE | 18 | 482 | 500 | |
| IME | 13 | 55 | 68 | |
| Relaxase | 50 | 13,066 | 13,116 | |
| Auxiliary protein | 44 | 7,614 | 7,658 | |
| T4CP | 16 | 16,969 | 16,985 | |
| Relaxosome | 6 | 15 | 21 | |
Usage
How to use oriTDB?

The web-based oriTDB database contains seven major parts: Home, Browse, Statistics, Tools, Download, References and Help. Users can browse and download resources according to their needs on the corresponding pages.
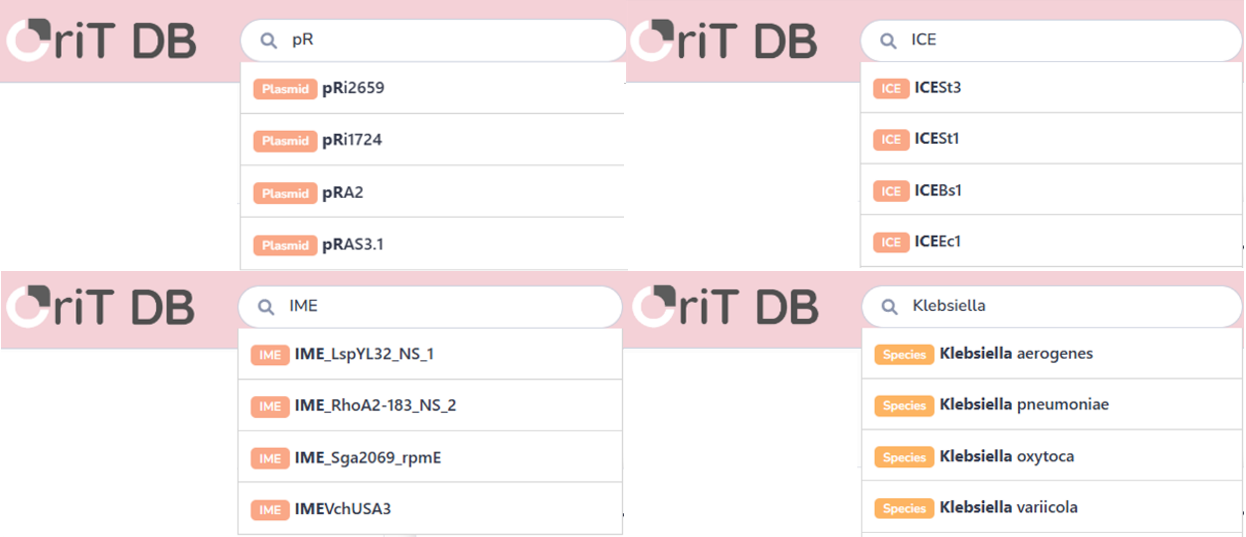
By entering a specific name of an MGE or a species in the search bar, users can quickly obtain detailed information about the oriT carried by a particular MGE or a category of species.
On the "Browse" page

The oriTDB database has classified the collected data on the oriT region, relaxase, auxiliary protein, T4CP, T4SS and relaxosome datasets into two major groups based on their sources (plasmids, ICEs, IMEs, vectors, and artificial transposons) and evidence (experimental validation and in silico prediction). Furthermore, users can conduct a fuzzy search using the search bar. Entries displayed in the browsing interface or search results can be selected to access the corresponding interface and obtain more detailed information.
On the "Statistics" page
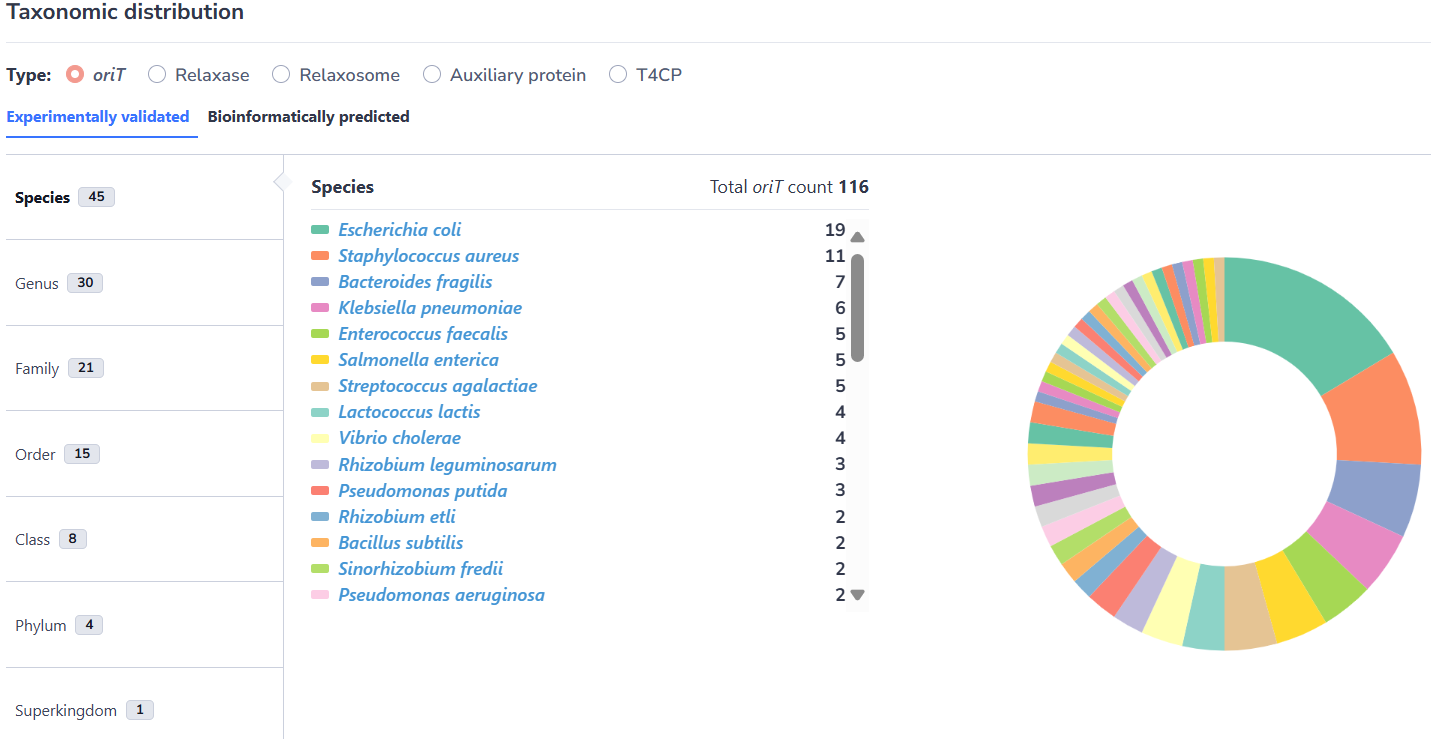
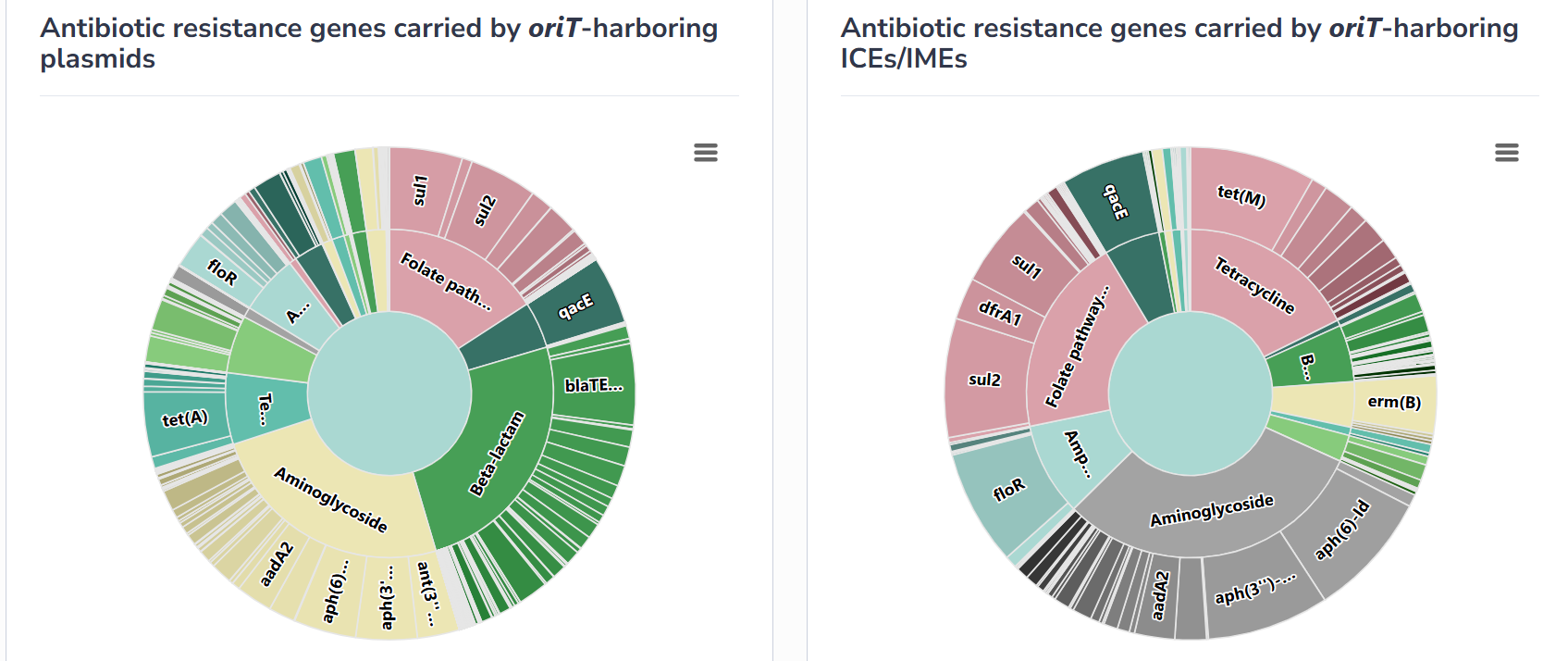
In this interface, the taxonomic distribution of the conjugative modules and the category distribution of cargo genes archived by oriTDB are displayed and illustrated through interactive pie charts. For the taxonomic distribution section, by clicking on the corresponding species in the pie chart, users can efficiently explore the MGEs of that species archived in oriTDB. For the cargo genes section, such as ARGs, by clicking on an ARG family in the pie chart, users can view in detail the distribution of ARG genes belong to this family. The same applies to other types of cargo genes, such as virulence factors, metal resistance genes and anti-CRISPRs.
On the "Tools" page
In this interface, oriTDB offers three online tools: oriTfinder2, BLAST search, and HMMER search:
On the "Download" page
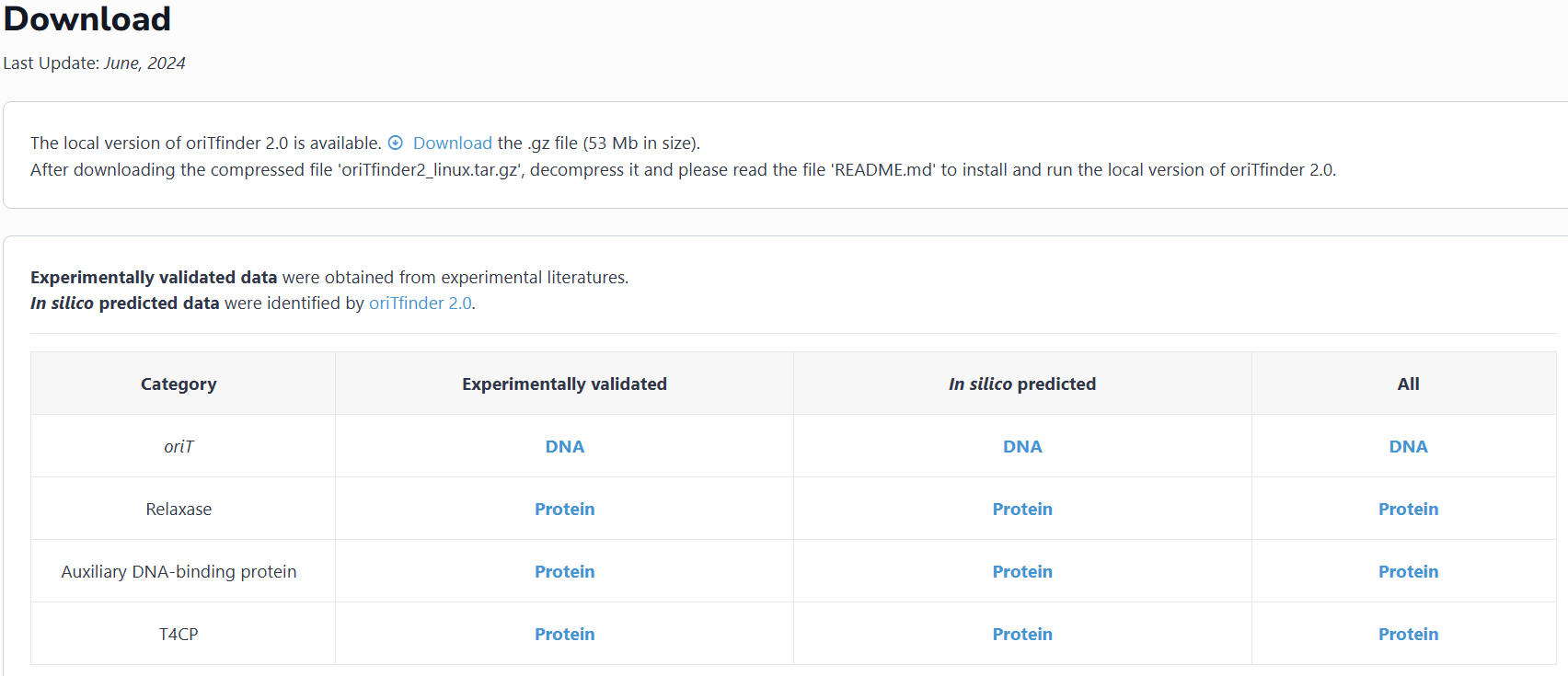
This interface provides the download for the local version of oriTfinder2. Users can also find all the datasets archived in oriTDB on this page and download them for free.
On the "References" page
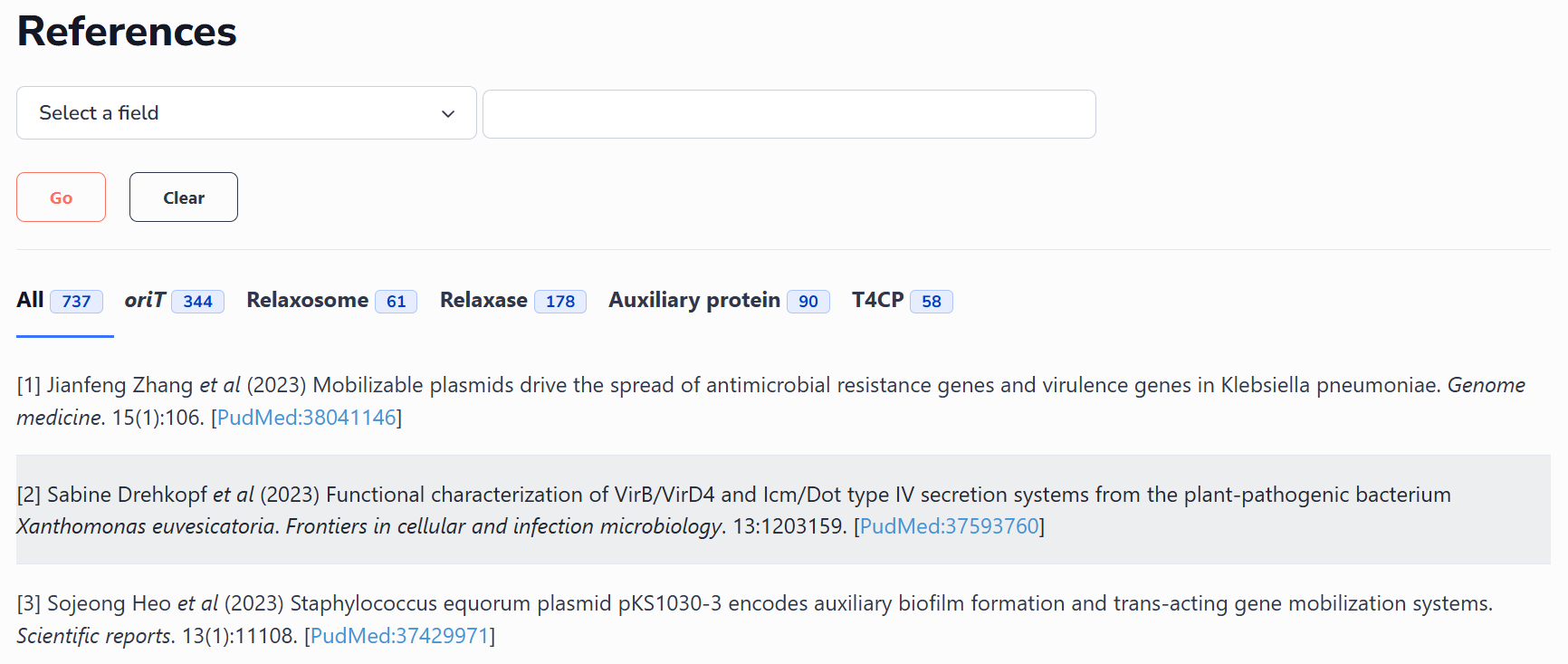

This interface includes all the oriT-related references collected by oriTDB and has classified them according to the conjugative modules (oriT region, relaxosome, relaxase, auxiliary protein, and T4CP). oriTDB also supports the search of included references using keywords such as author, article title, journal, year, and PubMed ID.
FAQs
Q1. How to use oriTDB?
A1: You can quickly understand the core functions of oriTDB through the browse interface, and get more detailed through this Help manual.
Q2. In the entries collected by oriTDB, some content is displayed as "-", what does this mean?
A2: If "-" appears in an experimentally validated entry, it indicates that we were unable to find corresponding data in the reference literature; if "-" appears in an in silico predicted entry, it means that the corresponding content was not predicted by oriTfinder.
Q3: How could I contact you if I find an error or have suggestions for oriTDB?
A3: Please do not hesitate to contact us through the mailing of hyou@sjtu.edu.cn.
Q4. Can I submit data to oriTDB?
A4: You can contact Prof. Ou for cooperation through the mailing of hyou@sjtu.edu.cn.