Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100003 |
| Name | oriT_RSF1010 |
| Organism | Escherichia coli |
| Sequence Completeness | core |
| NCBI accession of oriT (coordinates [strand]) | M28829 (3132..3169 [-], 38 nt) |
| oriT length | 38 nt |
| IRs (inverted repeats) | 1..10, 14..23 (CCAGTTTCTC..GAGAAACCGG) |
| Location of nic site | 31..32 |
| Conserved sequence flanking the nic site |
TAAGTGCG|CCCT |
| Note | _ |
oriT sequence
Download Length: 38 nt
CCAGTTTCTCGAAGAGAAACCGGTAAGTGCGCCCTCCC
Visualization of oriT structure (The oriT was characterized experimentally)
oriT secondary structure
Predicted by RNAfold.
Download structure file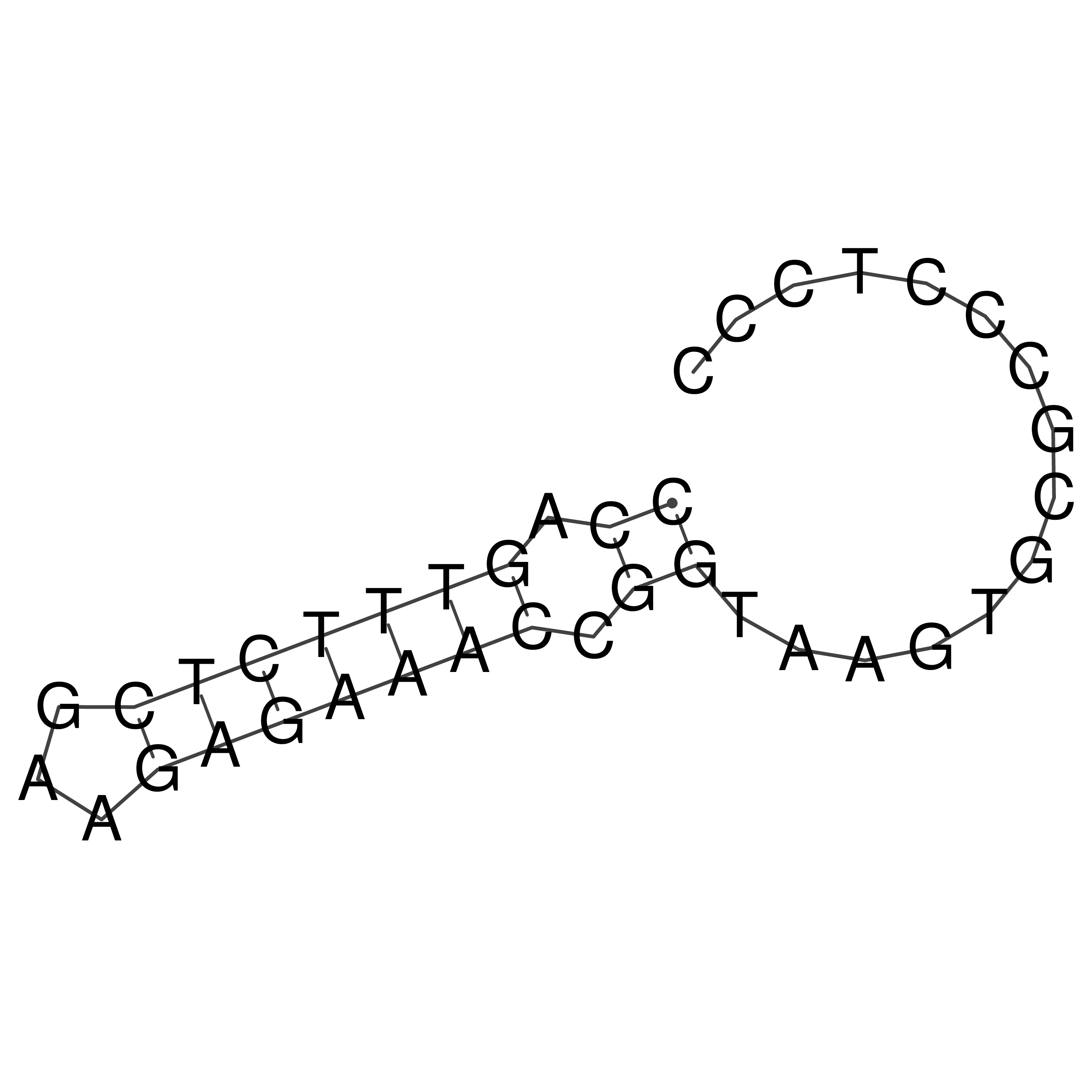
Reference
[1] Meyer R (2009) Replication and conjugative mobilization of broad host-range IncQ plasmids. Plasmid. 62(2):57-70. [PMID:19465049]
[2] Scherzinger E et al. (1993) Purification of the large mobilization protein of plasmid RSF1010 and characterization of its site-specific DNA-cleaving/DNA-joining activity. Eur J Biochem. 217(3):929-38. [PMID:8223650]
[3] Cook DM et al. (1992) The oriT region of the Agrobacterium tumefaciens Ti plasmid pTiC58 shares DNA sequence identity with the transfer origins of RSF1010 and RK2/RP4 and with T-region borders. J Bacteriol. 174(19):6238-46. [PMID:1400174]
Relaxosome
This oriT is a component of a relaxosome.
| Relaxosome name | RelaxosomeRSF1010 |
| oriT | oriT_RSF1010 |
| Relaxase | MobA_RSF1010 |
| Auxiliary protein | MobB_RSF1010 |
Reference
[1] Meyer R (2009) Replication and conjugative mobilization of broad host-range IncQ plasmids. Plasmid. 62(2):57-70. [PMID:19465049]
[2] Scherzinger E et al. (1993) Purification of the large mobilization protein of plasmid RSF1010 and characterization of its site-specific DNA-cleaving/DNA-joining activity. Eur J Biochem. 217(3):929-38. [PMID:8223650]
[3] Cook DM et al. (1992) The oriT region of the Agrobacterium tumefaciens Ti plasmid pTiC58 shares DNA sequence identity with the transfer origins of RSF1010 and RK2/RP4 and with T-region borders. J Bacteriol. 174(19):6238-46. [PMID:1400174]
Relaxase
| ID | 3 | GenBank | AAA26445 |
| Name | MobA_RSF1010 |
UniProt ID | P07112 |
| Length | 709 a.a. | PDB ID | |
| Note | relaxase | ||
Relaxase protein sequence
Download Length: 709 a.a. Molecular weight: 77951.91 Da Isoelectric Point: 7.2077
MAIYHLTAKTGSRSGGQSARAKADYIQREGKYARDMDEVLHAESGHMPEFVERPADYWDAADLYERANGR
LFKEVEFALPVELTLDQQKALASEFAQHLTGAERLPYTLAIHAGGGENPHCHLMISERINDGIERPAAQW
FKRYNGKTPEKGGAQKTEALKPKAWLEQTREAWADHANRALERAGHDARIDHRTLEAQGIERLPGVHLGP
NVVEMEGRGIRTDRADVALNIDTANAQIIDLQEYREAIDHERNRQSEEIQRHQRVSGADRTAGPEHGDTG
RRSPAGHEPDPAGQRGAGGGVAESPAPDRGGMGGAGQRVAGGSRRGEQRRAERPERVAGVALEAMANRDA
GFHDAYGGAADRIVALARPDATDNRGRLDLAALGGPMKNDRTLQAIGRQLKAMGCERFDIGVRDATTGQM
MNREWSAAEVLQNTPWLKRMNAQGNDVYIRPAEQERHGLVLVDDLSEFDLDDMKAEGREPALVVETSPKN
YQAWVKVADAAGGELRGQIARTLASEYDADPASADSRHYGRLAGFTNRKDKHTTRAGYQPWVLLRESKGK
TATAGPALVQQAGQQIEQAQRQQEKARRLASLELPERQLSRHRRTALDEYRSEMAGLVKRFGDDLSKCDF
IAAQKLASRGRSAEEIGKAMAEASPALAERKPGHEADYIERTVSKVMGLPSVQLARAELARAPAPRQRGM
DRGGPDFSM
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| PDB | 2NS6 | |
| AlphaFold DB | P07112 |
Reference
[1] Meyer R (2009) Replication and conjugative mobilization of broad host-range IncQ plasmids. Plasmid. 62(2):57-70. [PMID:19465049]
[2] Scherzinger E et al. (1993) Purification of the large mobilization protein of plasmid RSF1010 and characterization of its site-specific DNA-cleaving/DNA-joining activity. Eur J Biochem. 217(3):929-38. [PMID:8223650]
Auxiliary protein
| ID | 7 | GenBank | AAA26446 |
| Name | MobB_RSF1010 |
UniProt ID | P07113 |
| Length | 137 a.a. | PDB ID | _ |
| Note | _ | ||
Auxiliary protein sequence
Download Length: 137 a.a. Molecular weight: 15112.46 Da Isoelectric Point: 5.8900
MNAIDRVKKSRGINELAEQIEPLAQSMATLADEARQVMSQTQQASEAQAAEWLKAQRQTGAAWVELAKEL
REVAAEVSSAAQSARSASRGWHWKLWLTVMLASMMPTVVLLIASLLLLDLTPLTTEDGSIWLRLVAR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | P07113 |
Reference
[1] Scherzinger E et al. (1993) Purification of the large mobilization protein of plasmid RSF1010 and characterization of its site-specific DNA-cleaving/DNA-joining activity. Eur J Biochem. 217(3):929-38. [PMID:8223650]
| ID | 8 | GenBank | AAA26444 |
| Name | MobC_RSF1010 |
UniProt ID | P07114 |
| Length | 94 a.a. | PDB ID | _ |
| Note | _ | ||
Auxiliary protein sequence
Download Length: 94 a.a. Molecular weight: 10883.41 Da Isoelectric Point: 10.4580
MVKGSNKAADRLAKLEEQRARINAEIQRVRAREQQQERKNETRRKVLVGAMILAKVNSSEWPEDRLMAAM
DAYLERDHDRALFGLPPRQKDEPG
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | P07114 |
Reference
[1] Scherzinger E et al. (1993) Purification of the large mobilization protein of plasmid RSF1010 and characterization of its site-specific DNA-cleaving/DNA-joining activity. Eur J Biochem. 217(3):929-38. [PMID:8223650]
Host bacterium
| ID | 3 | GenBank | M28829 |
| Plasmid name | RSF1010 | Incompatibility group | IncQ1 |
| Plasmid size | 8684 bp | Coordinate of oriT [Strand] | 3132..3169 [-] |
| Host baterium | Escherichia coli |