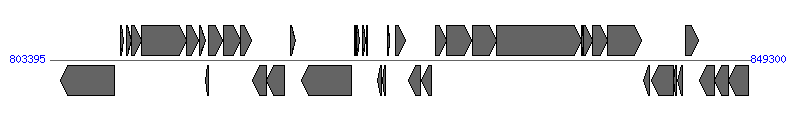The information of T4SS components from NC_010698 |
 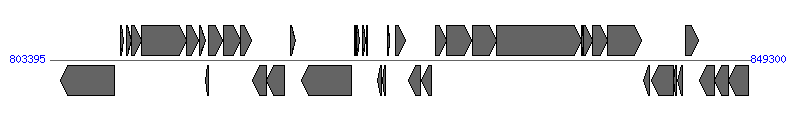 |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | HPSH_04145 | 804109..807594 [-], 3486 | 188527607 | cag pathogenicity island protein CagA | | |
| 2 | HPSH_04150 | 808001..808228 [+], 228 | 188527608 | cag pathogenicity island protein CagB | | |
| 3 | HPSH_04155 | 808395..808742 [+], 348 | 188527609 | cag pathogenicity island protein CagC | CagC | |
| 4 | HPSH_04160 | 808744..809367 [+], 624 | 188527610 | cag pathogenicity island protein CagD | CagD | |
| 5 | HPSH_04165 | 809382..812327 [+], 2946 | 188527611 | cag pathogenicity island protein CagE | CagE | |
| 6 | HPSH_04170 | 812329..813135 [+], 807 | 188527612 | cag pathogenicity island protein CagF | CagF |  |
| 7 | HPSH_04175 | 813190..813621 [+], 432 | 188527613 | cag pathogenicity island protein CagG | CagG | |
| 8 | HPSH_04180 | 813607..813804 [-], 198 | 188527614 | hypothetical protein | | |
| 9 | HPSH_04185 | 813803..814735 [+], 933 | 188527615 | cag pathogenicity island protein CagH | CagH | |
| 10 | HPSH_04190 | 814746..815885 [+], 1140 | 188527616 | cag pathogenicity island protein CagI | CagI | |
| 11 | HPSH_04195 | 815882..816595 [+], 714 | 188527617 | cag pathogenicity island protein CagL | CagL |  |
| 12 | HPSH_04200 | 816678..817598 [-], 921 | 188527618 | cag pathogenicity island protein CagN | CagN | |
| 13 | HPSH_04205 | 817613..818743 [-], 1131 | 188527619 | cag pathogenicity island protein CagM | CagM | |
| 14 | HPSH_04210 | 819169..819513 [+], 345 | 188527620 | cag pathogenicity island protein CagP | | |
| 15 | HPSH_04215 | 819912..823169 [-], 3258 | 188527621 | cag pathogenicity island protein CagA | | |
| 16 | HPSH_04220 | 823362..823460 [+], 99 | 188527622 | hypothetical protein | | |
| 17 | HPSH_04225 | 823481..823708 [+], 228 | 188527623 | cag pathogenicity island protein CagB | | |
| 18 | HPSH_04230 | 823876..824067 [+], 192 | 188527624 | cag pathogenicity island protein CagC | CagC | |
| 19 | HPSH_04235 | 824137..824232 [+], 96 | 188527625 | hypothetical protein | | |
| 20 | HPSH_04240 | 824858..825163 [-], 306 | 188527626 | cag pathogenicity island protein CagQ | | |
| 21 | HPSH_04245 | 825192..825374 [-], 183 | 188527627 | hypothetical protein | | |
| 22 | HPSH_04250 | 825507..825644 [+], 138 | 188527628 | hypothetical protein | | |
| 23 | HPSH_04255 | 826078..826677 [+], 600 | 188527629 | cag pathogenicity island protein CagS | | |
| 24 | HPSH_04260 | 826879..827721 [-], 843 | 188527630 | cag pathogenicity island protein CagT | CagT | |
| 25 | HPSH_04265 | 827737..828384 [-], 648 | 188527631 | cag pathogenicity island protein CagU | CagU | |
| 26 | HPSH_04270 | 828660..829418 [+], 759 | 188527632 | cag pathogenicity island protein CagV | CagV | |
| 27 | HPSH_04275 | 829423..831030 [+], 1608 | 188527633 | cag pathogenicity island protein CagW | CagW | |
| 28 | HPSH_04280 | 831082..832650 [+], 1569 | 188527634 | cag pathogenicity island protein CagX | CagX | |
| 29 | HPSH_04285 | 832665..838229 [+], 5565 | 188527635 | cag pathogenicity island protein CagY | CagY | |
| 30 | HPSH_04290 | 838219..838353 [+], 135 | 188527636 | hypothetical protein | | |
| 31 | HPSH_04295 | 838366..838965 [+], 600 | 188527637 | cag pathogenicity island protein CagZ | CagZ | |
| 32 | HPSH_04300 | 838970..839962 [+], 993 | 188527638 | cag pathogenicity island protein cag alpha | Cagalpha | |
| 33 | HPSH_04305 | 839971..842217 [+], 2247 | 188527639 | cag pathogenicity island protein Cag beta | Cagbeta | |
| 34 | HPSH_04310 | 842336..842716 [-], 381 | 188527640 | cag pathogenicity island protein Cag gamma | Caggamma | |
| 35 | HPSH_04315 | 842855..844300 [-], 1446 | 188527641 | cag pathogenicity island protein Cag delta | Cagdelta | |
| 36 | HPSH_04320 | 844314..844511 [-], 198 | 188527642 | cag pathogenicity island protein Cag theta | | |
| 37 | HPSH_04325 | 844527..844874 [-], 348 | 188527643 | cag pathogenicity island protein Cag zeta | | |
| 38 | HPSH_04330 | 845087..845917 [+], 831 | 188527644 | hypothetical protein | | |
| 39 | HPSH_04335 | 845992..846984 [-], 993 | 188527645 | hypothetical protein | | |
| 40 | HPSH_04340 (era) | 846981..847886 [-], 906 | 188527646 | GTP-binding protein Era | | |
| 41 | HPSH_04345 (hslU) | 847886..849217 [-], 1332 | 188527647 | ATP-dependent protease ATP-binding subunit HslU | | |
| |