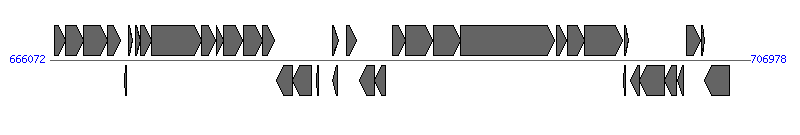The information of T4SS components from NC_014256 |
 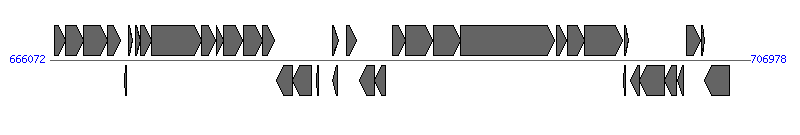 |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | HPB8_691 (nudT14) | 666318..666956 [+], 639 | 298736186 | UDP-sugar diphosphatase | | |
| 2 | HPB8_692 | 666967..668022 [+], 1056 | 298736187 | plasminogen-binding protein PgbA | | |
| 3 | HPB8_693 (glcD1) | 668024..669403 [+], 1380 | 298736188 | FAD linked oxidase-like protein | | |
| 4 | HPB8_694 (dapB) | 669425..670189 [+], 765 | 298736189 | Dihydrodipicolinate reductase | | |
| 5 | HPB8_695 | 670413..670523 [-], 111 | 298736190 | hypothetical protein | | |
| 6 | HPB8_696 (cagB) | 670637..670864 [+], 228 | 298736191 | cag pathogenicity island protein B | | |
| 7 | HPB8_697 (cagC) | 671072..671356 [+], 285 | 298736192 | cag pathogenicity island protein C | CagC | |
| 8 | HPB8_698 (cagD) | 671340..671984 [+], 645 | 298736193 | cag pathogenicity island protein D | CagD | |
| 9 | HPB8_699 (cagE) | 671999..674944 [+], 2946 | 298736194 | cag pathogenicity island protein E | CagE | |
| 10 | HPB8_700 (cagF) | 674946..675752 [+], 807 | 298736195 | cag pathogenicity island protein F | CagF |  |
| 11 | HPB8_701 (cagG) | 675807..676235 [+], 429 | 298736196 | cag pathogenicity island protein G | CagG | |
| 12 | HPB8_702 (cagH) | 676237..677349 [+], 1113 | 298736197 | cag pathogenicity island protein H | CagH | |
| 13 | HPB8_703 (cagI) | 677360..678505 [+], 1146 | 298736198 | cag pathogenicity island protein I | CagI | |
| 14 | HPB8_704 (cagL) | 678502..679215 [+], 714 | 298736199 | cag pathogenicity island protein L | CagL |  |
| 15 | HPB8_705 (cagN) | 679297..680223 [-], 927 | 298736200 | cag pathogenicity island protein N | CagN | |
| 16 | HPB8_706 (cagM) | 680232..681362 [-], 1131 | 298736201 | cag pathogenicity island protein M | CagM | |
| 17 | HPB8_707 | 681666..681782 [-], 117 | 298736202 | hypothetical protein | | |
| 18 | HPB8_708 | 682552..682854 [-], 303 | 298736203 | hypothetical protein | | |
| 19 | HPB8_709 (cagQ) | 682562..682942 [+], 381 | 298736204 | cag pathogenicity island protein Q | | |
| 20 | HPB8_710 (cagS) | 683384..683983 [+], 600 | 298736205 | cag pathogenicity island protein S | | |
| 21 | HPB8_711 (cagT) | 684180..685022 [-], 843 | 298736206 | cag pathogenicity island protein T | CagT | |
| 22 | HPB8_712 (cagU) | 685035..685685 [-], 651 | 298736207 | cag pathogenicity island protein U | CagU | |
| 23 | HPB8_713 (cagV) | 686072..686830 [+], 759 | 298736208 | cag pathogenicity island protein V | CagV | |
| 24 | HPB8_714 (cagW) | 686835..688433 [+], 1599 | 298736209 | cag pathogenicity island protein W | CagW | |
| 25 | HPB8_715 (cagX) | 688486..690054 [+], 1569 | 298736210 | cag pathogenicity island protein X | CagX | |
| 26 | HPB8_716 (cagY) | 690069..695534 [+], 5466 | 298736211 | cag pathogenicity island protein Y | CagY | |
| 27 | HPB8_717 (cagZ) | 695683..696282 [+], 600 | 298736212 | cag pathogenicity island protein Z | CagZ | |
| 28 | HPB8_718 (virB11-4) | 696287..697279 [+], 993 | 298736213 | cag pathogenicity island protein alpha | Cagalpha | |
| 29 | HPB8_719 (cag5) | 697288..699534 [+], 2247 | 298736214 | cag pathogenicity island protein beta | Cagbeta | |
| 30 | HPB8_720 | 699579..699698 [-], 120 | 298736215 | hypothetical protein | | |
| 31 | HPB8_721 | 699618..699893 [+], 276 | 298736216 | hypothetical protein | | |
| 32 | HPB8_722 (cag4) | 700014..700523 [-], 510 | 298736217 | cag pathogenicity island protein gamma | Caggamma | |
| 33 | HPB8_723 (cag3) | 700533..701978 [-], 1446 | 298736218 | cag pathogenicity island protein delta | Cagdelta | |
| 34 | HPB8_724 (cag2) | 701971..702675 [-], 705 | 298736219 | cag pathogenicity island protein epsilon | | |
| 35 | HPB8_725 (cag1) | 702739..703086 [-], 348 | 298736220 | cag pathogenicity island protein zeta | | |
| 36 | HPB8_726 | 703259..704110 [+], 852 | 298736221 | chitin synthase regulatory factor 3 | | |
| 37 | HPB8_727 | 704147..704326 [+], 180 | 298736222 | hypothetical protein | | |
| 38 | HPB8_728 (glnA) | 704340..705785 [-], 1446 | 298736223 | glutamine synthetase | | |
| |