Introduction
TADB 3.0 is a comprehensive database that dedicated to unraveling the intricate relationship between bacterial Toxin-Antitoxin (TA) systems and Mobile Genetic Elements (MGEs).
TADB 3.0 represents a significant improvement over previous versions, which offers three major enhancements: (i) manual curation of > 500 types I to VIII TA loci with experimental evidence and support; (ii) collection of the TAfinder 2.0-predicted TA loci in > 34,000 completely sequenced prokaryotic genomes; (iii) graphical representation of the relationships of the TA loci and MGEs.
It will facilitate research on in silico prediction of TA systems and research on the interplay between TA systems and MGEs.
TA system dataset
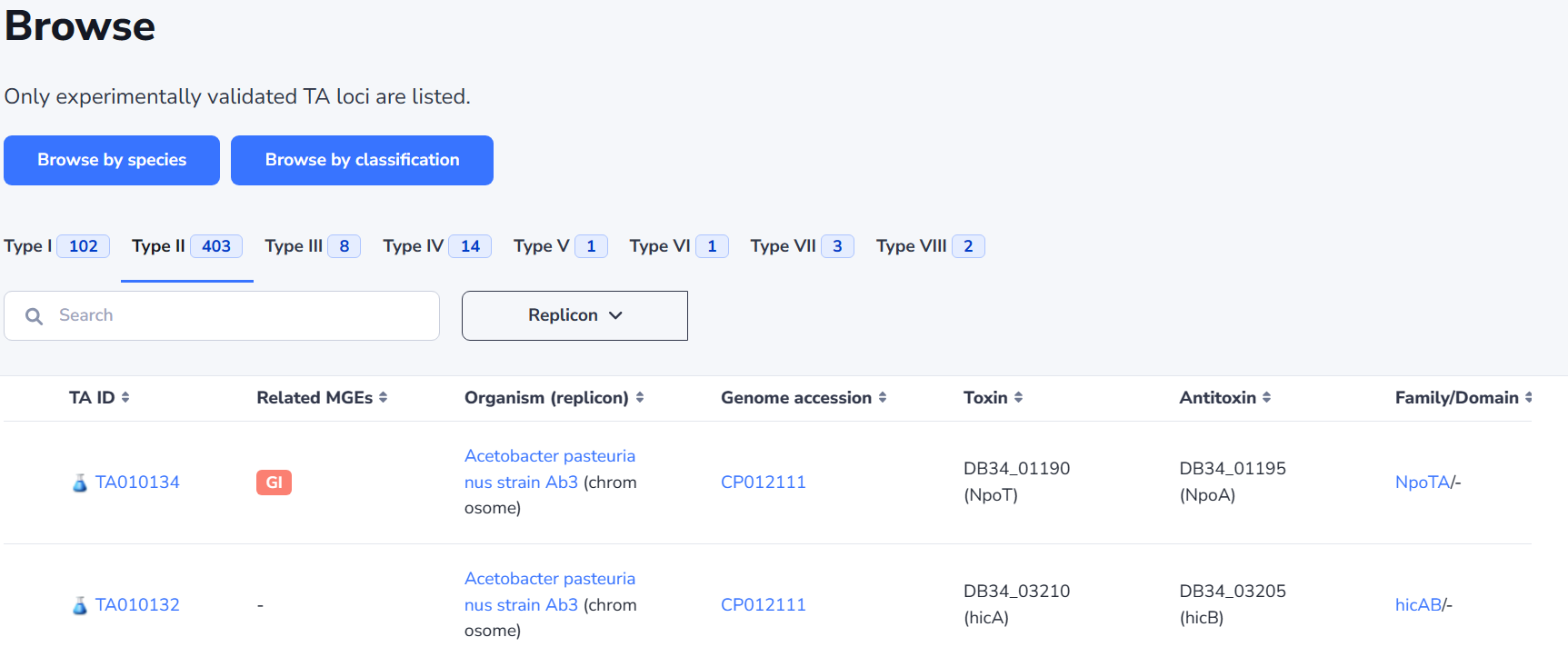
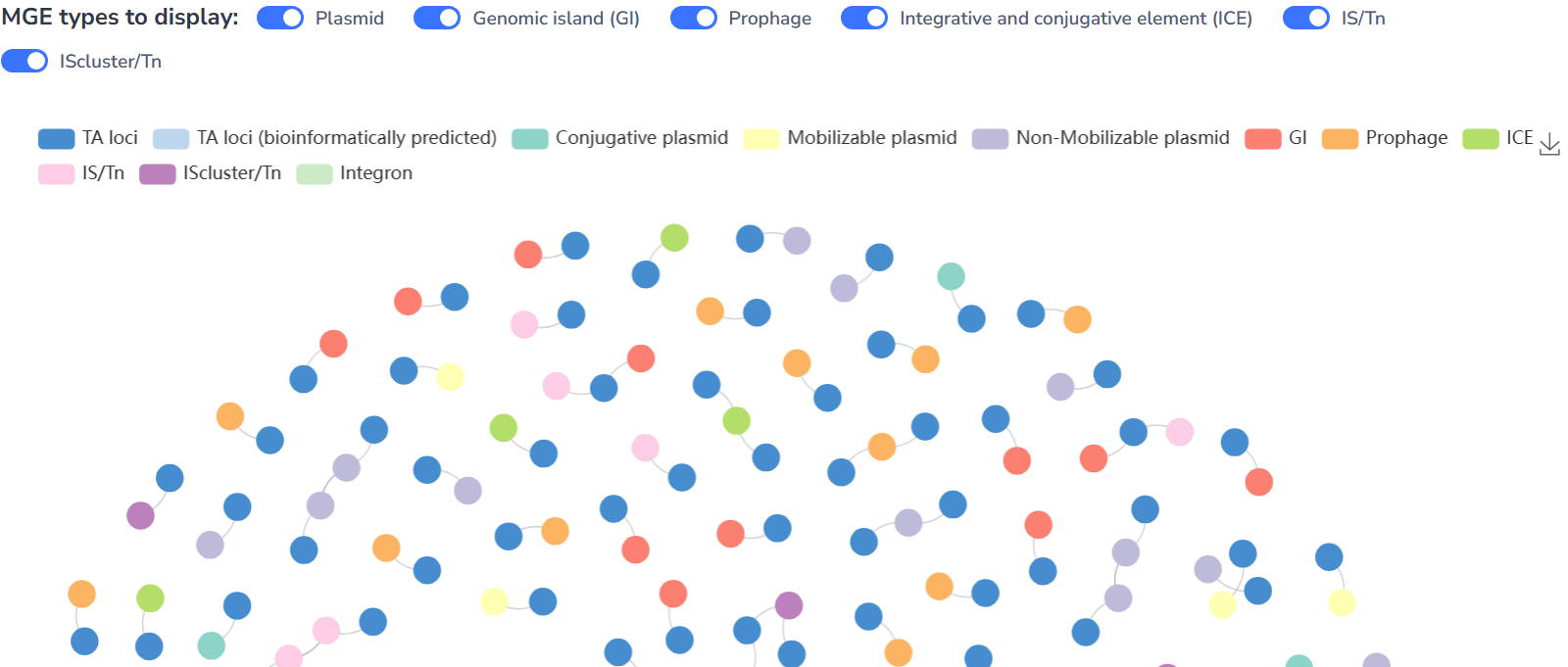
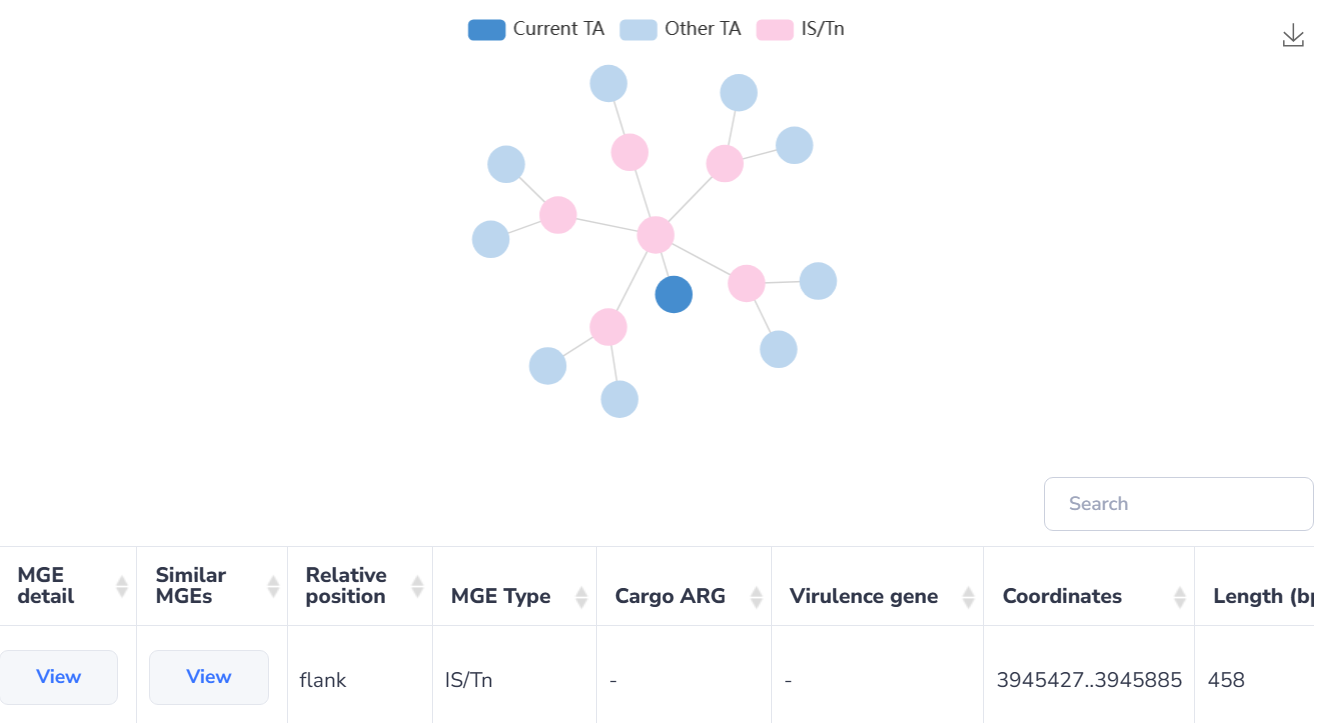
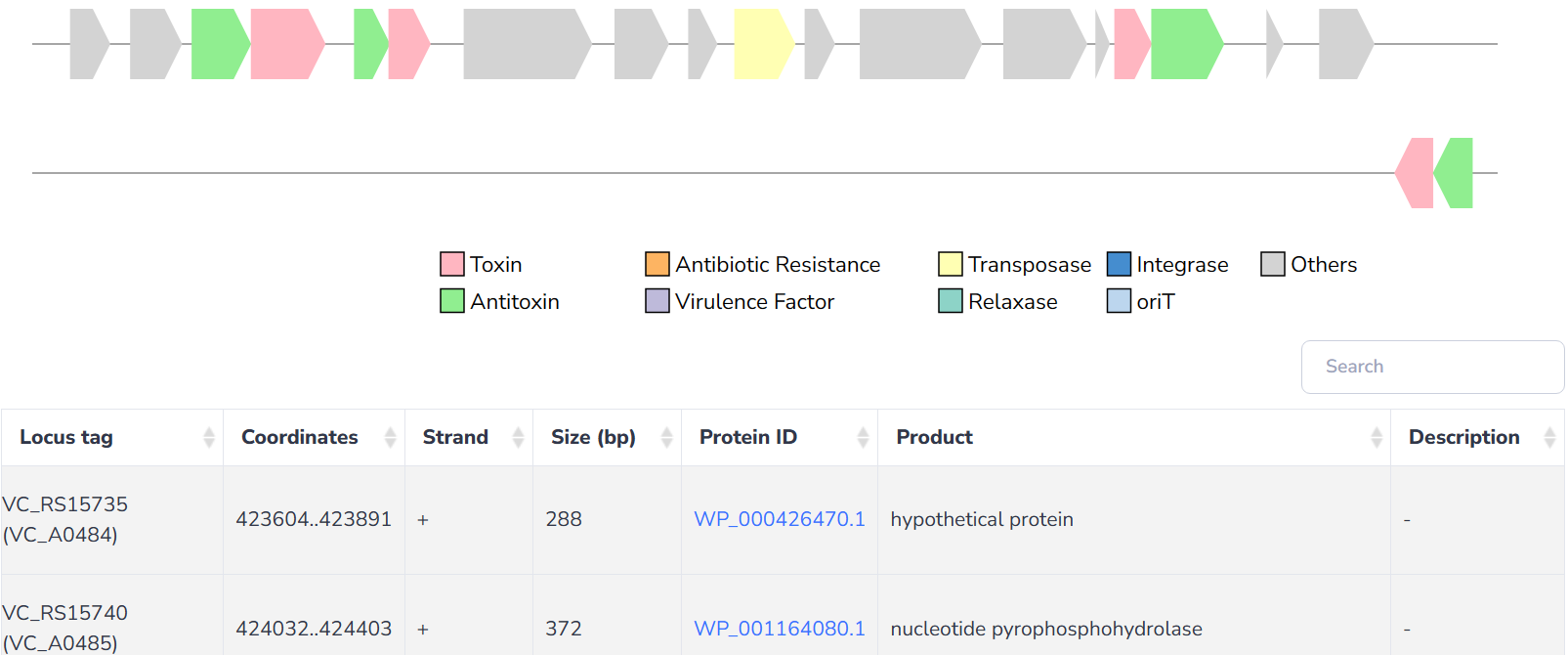
TA-MGE relationships
TA systems are found to be passengers on almost all kinds of MGEs, which allow these systems to rapidly disseminate through bacterial communities by horizontal transfer. Moreover, TA systems are suggested to contribute to the maintenance of MGEs, ensuring their persistence in bacterial populations and contributing to the survival of bacteria under environmental stress.
TAfinder 2.0
By using the newly added experimentally validated TA loci data, TAfinder is now able to identify all types of TA system in annotated or unannotated bacterial genomes. The workflow of TAfinder2 are described below:
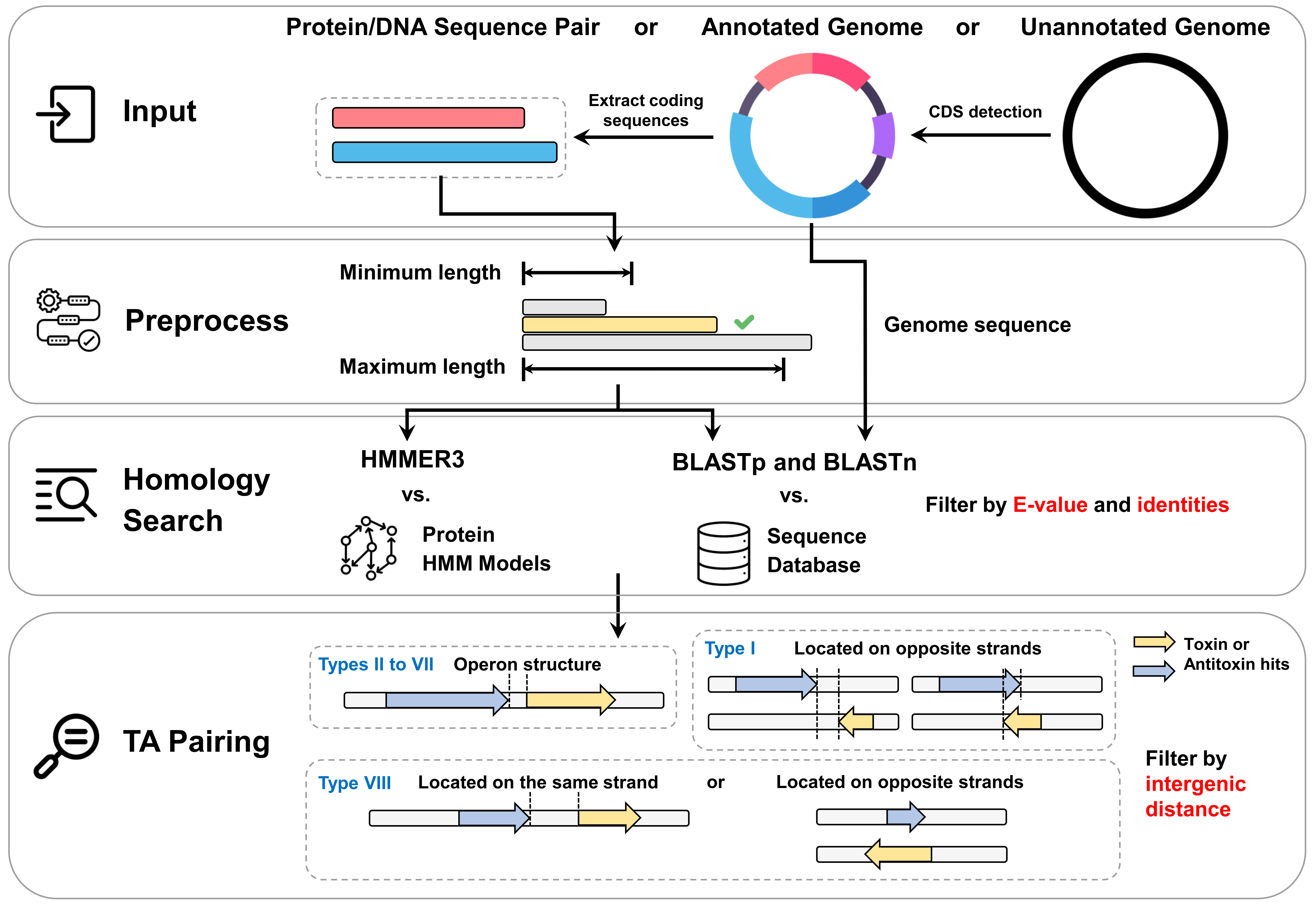
For the input section, three types of input are acceptable: a pair of protein or DNA sequences, an annotated genome in the GenBank format, and an unannotated genome sequence in the FASTA format. For an unannotated genome sequence input,
Prodigal would be used for protein-coding sequence (CDS) identification before sequence extraction. In the preprocess section, the extracted sequences would be filtered by user-defined maximum length (500 a.a. by default) and minimum length (30 a.a. by default).
In the homology search section, the protein sequences are input to BLASTp and HMMER3 to search for protein homologues, while the genome sequence is input to BLASTn to identify RNA toxins and antitoxins. The E-value (0.01 by default) for BLAST and HMMER3 as well as the identities (30% by default) for BLAST are set to filter out the results.
In the TA pairing section, for the identification of types II to VII TA loci, the toxin hits and antitoxin hits should be located in the same strand, and the maximum intergenic distance (150 bp by default) is set for identifying the TA operon structure.
For the identification of type I TA loci, the toxin gene and antitoxin RNA should be located on the opposite DNA strands, rather than forming an operon structure on the same strand. In addition, for the identification of type VIII TA loci, we took into consideration the two experimentally validated type VIII systems.
One type VIII TA locus had both RNAs located on the same strand (Ming Li et al.), while the other locus had the two RNAs located on the opposite strands (Jee Soo Choi et al.). Consequently, we predicted these two distinct type VIII TA loci based on their respective characteristics.