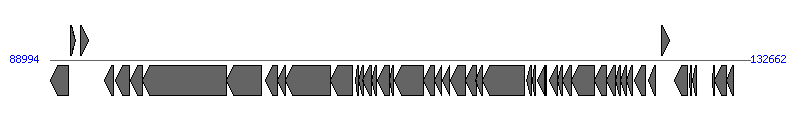The information of T4SS components from NC_010409 |
 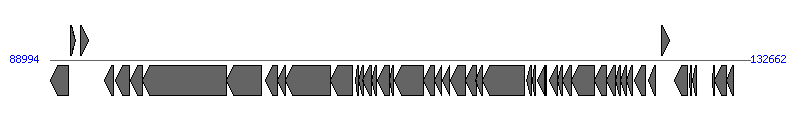 |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | pVM01_p080 | 89017..90150 [-], 1134 | 169546468 | hypothetical protein | | |
| 2 | pVM01_p081 | 90256..90579 [+], 324 | 169546507 | hypothetical protein | | |
| 3 | pVM01_p082 | 90875..91378 [+], 504 | 169546469 | hypothetical protein | | |
| 4 | pVM01_p083 | 92412..92972 [-], 561 | 169546470 | conjugal transfer fertility inhibition protein FinO | | |
| 5 | pVM01_p084 | 93075..93935 [-], 861 | 169546508 | hypothetical protein | | |
| 6 | pVM01_p085 | 93994..94740 [-], 747 | 169546471 | conjugal transfer pilus acetylation protein TraX | TraX_F | |
| 7 | pVM01_p086 | 94760..100030 [-], 5271 | 169546472 | conjugal transfer nickase/helicase TraI | TraI_F | |
| 8 | pVM01_p087 | 100030..102183 [-], 2154 | 169546473 | conjugal transfer protein TraD | TraD_F | |
| 9 | pVM01_p088 | 102436..103167 [-], 732 | 169546474 | conjugal transfer surface exclusion protein TraT | | |
| 10 | pVM01_p089 | 103181..103690 [-], 510 | 169546475 | conjugal transfer entry exclusion protein TraS | | |
| 11 | pVM01_p090 | 103687..106512 [-], 2826 | 169546476 | conjugal transfer mating pair stabilization protein TraG | TraG_F | |
| 12 | pVM01_p091 | 106509..107882 [-], 1374 | 169546477 | conjugal transfer pilus assembly protein TraH | TraH_F | |
| 13 | pVM01_p092 | 108073..108261 [-], 189 | 169546509 | hypothetical protein | | |
| 14 | pVM01_p093 | 108248..108529 [-], 282 | 169546478 | conjugal transfer protein TrbJ | | |
| 15 | pVM01_p094 | 108519..109064 [-], 546 | 169546479 | conjugal transfer protein TrbB | | |
| 16 | pVM01_p095 | 109051..109332 [-], 282 | 169546480 | conjugal transfer pilin chaperone TraQ | TraQ_F | |
| 17 | pVM01_p096 | 109448..110191 [-], 744 | 169546481 | conjugal pilus assembly protein TraF | TraF_F | |
| 18 | pVM01_p097 | 110184..110441 [-], 258 | 169546482 | conjugal transfer protein TrbE | | |
| 19 | pVM01_p098 | 110468..112318 [-], 1851 | 169546483 | conjugal transfer mating pair stabilization protein TraN | TraN_F | |
| 20 | pVM01_p099 | 112315..112953 [-], 639 | 169546484 | conjugal transfer pilus assembly protein TrbC | TrbC_F | |
| 21 | pVM01_p100 | 112981..113433 [-], 453 | 169546510 | hypothetical protein | | |
| 22 | pVM01_p101 | 113463..113924 [-], 462 | 169546511 | hypothetical protein | | |
| 23 | pVM01_p102 | 113950..114942 [-], 993 | 169546512 | conjugal transfer pilus assembly protein TraU | TraU_F | |
| 24 | pVM01_p103 | 114939..115571 [-], 633 | 169546513 | conjugal transfer pilus assembly protein TraW | TraW_F | |
| 25 | pVM01_p104 | 115568..115954 [-], 387 | 169546514 | conjugal transfer protein TrbI | | |
| 26 | pVM01_p105 | 115951..118581 [-], 2631 | 169546515 | conjugal transfer ATP-binding protein TraC | TraC_F | |
| 27 | pVM01_p106 | 118708..119055 [-], 348 | 169546516 | hypothetical protein | | |
| 28 | pVM01_p107 | 119083..119301 [-], 219 | 169546517 | hypothetical protein | | |
| 29 | pVM01_p108 | 119381..119854 [-], 474 | 169546518 | hypothetical protein | | |
| 30 | pVM01_p109 | 119903..119986 [-], 84 | 169546519 | hypothetical protein | | |
| 31 | pVM01_p110 | 120139..120654 [-], 516 | 169546520 | conjugal transfer protein TraV | TraV_F | |
| 32 | pVM01_p111 | 120651..120971 [-], 321 | 169546521 | conjugal transfer protein TrbD | | |
| 33 | pVM01_p112 | 120958..121548 [-], 591 | 169546522 | conjugal transfer protein TraP | | |
| 34 | pVM01_p113 | 121538..122965 [-], 1428 | 169546523 | conjugal transfer pilus assembly protein TraB | TraB_F | |
| 35 | pVM01_p114 | 122965..123693 [-], 729 | 169546524 | conjugal transfer protein TraK | TraK_F | |
| 36 | pVM01_p115 | 123680..124246 [-], 567 | 169546525 | conjugal transfer pilus assembly protein TraE | TraE_F | |
| 37 | pVM01_p116 | 124268..124579 [-], 312 | 169546526 | conjugal transfer pilus assembly protein TraL | TraL_F | |
| 38 | pVM01_p117 | 124594..124953 [-], 360 | 169546527 | conjugal transfer pilin subunit TraA | TraA_F | |
| 39 | pVM01_p118 | 124986..125345 [-], 360 | 169546528 | conjugal transfer protein TraY | TraY_F | |
| 40 | pVM01_p119 | 125480..126169 [-], 690 | 169546529 | conjugal transfer transcriptional regulator TraJ | | |
| 41 | pVM01_p120 | 126356..126739 [-], 384 | 169546530 | conjugal transfer protein TraM | TraM_F |  |
| 42 | pVM01_p121 | 127153..127662 [+], 510 | 169546531 | lytic transglycosylase | Orf169_F | |
| 43 | pVM01_p122 | 127959..128780 [-], 822 | 169546532 | hypothetical protein | | |
| 44 | pVM01_p123 | 128900..129040 [-], 141 | 169546533 | hypothetical protein | | |
| 45 | pVM01_p124 | 129075..129329 [-], 255 | 169546534 | hypothetical protein | | |
| 46 | pVM01_p125 | 130321..130458 [-], 138 | 169546535 | hypothetical protein | | |
| 47 | pVM01_p126 | 130458..131177 [-], 720 | 169546536 | plasmid SOS inhibition protein A | | |
| 48 | pVM01_p127 (psiB) | 131174..131608 [-], 435 | 169546537 | plasmid SOS inhibition protein B | | |
| |