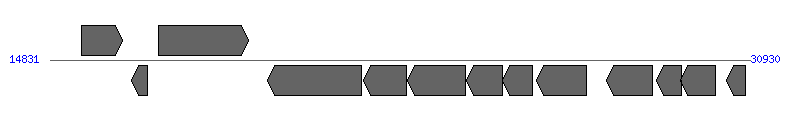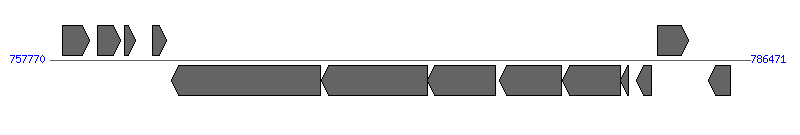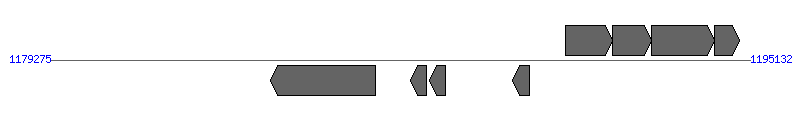The information of T4SS components from NC_007354 |
| Region 1: 19831..25930 |
 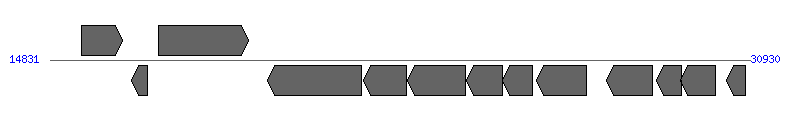 |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | Ecaj_0015 | 15553..16494 [+], 942 | 73666648 | quinolinate synthetase | | |
| 2 | Ecaj_0016 | 16694..17071 [-], 378 | 73666649 | 4Fe-4S ferredoxin | | |
| 3 | Ecaj_0017 | 17335..19401 [+], 2067 | 73666650 | gp140 | | |
| 4 | Ecaj_0018 | 19831..22002 [-], 2172 | 73666651 | type IV secretion system protein VirB8 | VirD4 | |
| 5 | Ecaj_0019 | 22041..23039 [-], 999 | 73666652 | type IV secretion system ATPase VirB11 | VirB11 | |
| 6 | Ecaj_0020 | 23056..24393 [-], 1338 | 73666653 | conjugation TrbI-like protein | VirB10 | |
| 7 | Ecaj_0021 | 24415..25239 [-], 825 | 73666654 | conjugal transfer protein TrbG/VirB9/CagX | VirB9 | |
| 8 | Ecaj_0022 | 25226..25930 [-], 705 | 73666655 | virulence protein | VirB8 | |
| 9 | Ecaj_0023 | 26023..27165 [-], 1143 | 73666656 | GTP cyclohydrolase II | | |
| 10 | Ecaj_0024 | 27624..28694 [-], 1071 | 73666657 | hypothetical protein | | |
| 11 | Ecaj_0025 | 28769..29353 [-], 585 | 73666658 | hypothetical protein | | |
| 12 | Ecaj_0026 | 29334..30137 [-], 804 | 73666659 | diaminopimelate epimerase | | |
| 13 | Ecaj_0027 | 30388..30819 [-], 432 | 73666660 | hypothetical protein | | |
| |
| Region 2: 762770..781471 |
 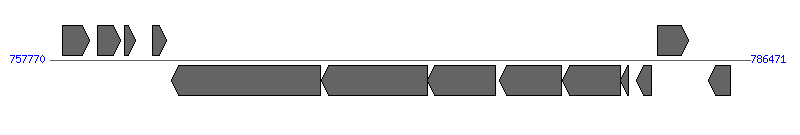 |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | Ecaj_0525 | 758290..759405 [+], 1116 | 73667143 | carbamoyl phosphate synthase small subunit | | |
| 2 | Ecaj_0526 (ispH) | 759707..760666 [+], 960 | 73667144 | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase | | |
| 3 | Ecaj_0527 | 760813..761274 [+], 462 | 73667145 | deoxyuridine 5'-triphosphate nucleotidohydrolase | | |
| 4 | Ecaj_0528 | 761965..762564 [+], 600 | 73667146 | hypothetical protein | | |
| 5 | Ecaj_0529 | 762770..768862 [-], 6093 | 73667147 | putative Type IV secretory pathway VirB6 components | VirB6 | |
| 6 | Ecaj_0530 | 768899..773233 [-], 4335 | 73667148 | TrbL/VirB6 plasmid conjugal transfer protein | VirB6 | |
| 7 | Ecaj_0531 | 773237..776044 [-], 2808 | 73667149 | TrbL/VirB6 plasmid conjugal transfer protein | VirB6 | |
| 8 | Ecaj_0532 | 776207..778726 [-], 2520 | 73667150 | TrbL/VirB6 plasmid conjugal transfer protein | VirB6 | |
| 9 | Ecaj_0533 | 778764..781166 [-], 2403 | 73667151 | type IV secretion system ATPase VirB4 | VirB4 | |
| 10 | Ecaj_0534 | 781166..781471 [-], 306 | 73667152 | type IV secretion system protein VirB3 | VirB3 | |
| 11 | Ecaj_0535 | 781835..782452 [-], 618 | 73667153 | superoxide dismutase | | |
| 12 | Ecaj_0536 | 782681..783946 [+], 1266 | 73667154 | binding-protein dependent transport system inner membrane protein | | |
| 13 | Ecaj_0537 | 784752..785651 [-], 900 | 73667155 | lipoyl synthase | | |
| |
| Region 3: 1104922..1105734 |
 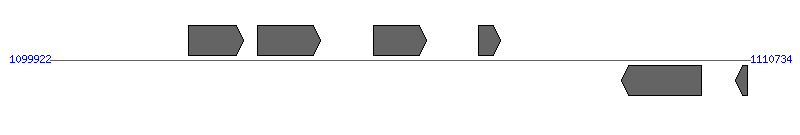 |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | Ecaj_0785 | 1102054..1102905 [+], 852 | 73667398 | Alpha/beta hydrolase | | |
| 2 | Ecaj_0786 | 1103119..1104102 [+], 984 | 73667399 | pyruvate dehydrogenase (lipoamide) | | |
| 3 | Ecaj_0787 | 1104922..1105734 [+], 813 | 73667400 | conjugal transfer protein TrbG/VirB9/CagX | VirB9 | |
| 4 | Ecaj_0788 | 1106543..1106875 [+], 333 | 73667401 | thioredoxin | | |
| 5 | Ecaj_0789 | 1108752..1109987 [-], 1236 | 73667402 | tRNA 2-methylthioadenosine synthase | | |
| 6 | Ecaj_0790 | 1110510..1110698 [-], 189 | 73667403 | exodeoxyribonuclease VII small subunit | | |
| |
| Region 4: 1184275..1190132 |
 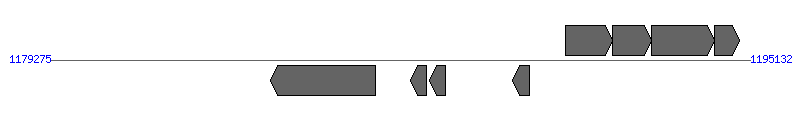 |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | Ecaj_0839 | 1184275..1186650 [-], 2376 | 73667452 | type IV secretion system protein VirB5 | VirB4 | |
| 2 | Ecaj_0840 | 1187439..1187801 [-], 363 | 73667453 | hypothetical protein | VirB2 | |
| 3 | Ecaj_0841 | 1187873..1188238 [-], 366 | 73667454 | hypothetical protein | VirB2 | |
| 4 | Ecaj_0842 | 1189755..1190132 [-], 378 | 73667455 | hypothetical protein | VirB2 | |
| 5 | Ecaj_0843 | 1190954..1192021 [+], 1068 | 73667456 | HflK protein | | |
| 6 | Ecaj_0844 | 1192026..1192898 [+], 873 | 73667457 | hypothetical protein | | |
| 7 | Ecaj_0845 | 1192902..1194317 [+], 1416 | 73667458 | peptidase S1 | | |
| 8 | Ecaj_0846 | 1194319..1194900 [+], 582 | 73667459 | hypothetical protein | | |
| |