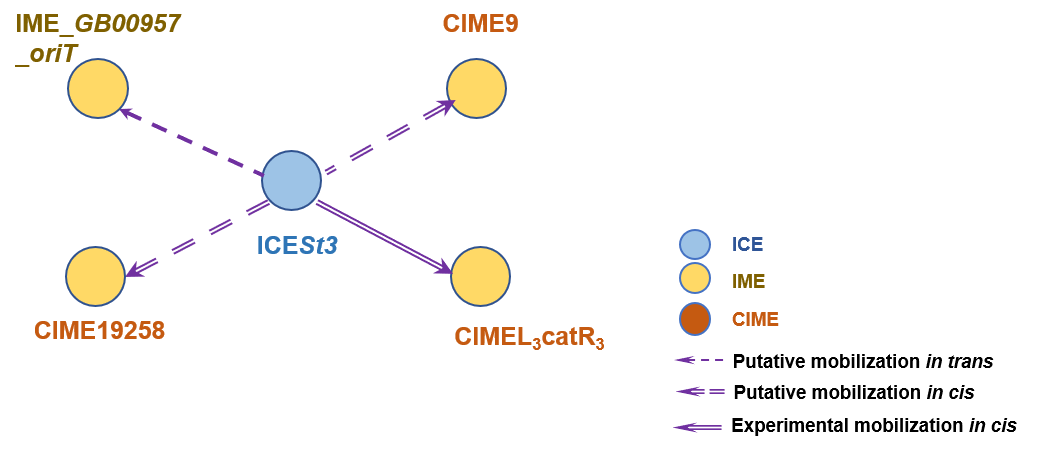
The graph information of ICESt3 components from AJ586568 |
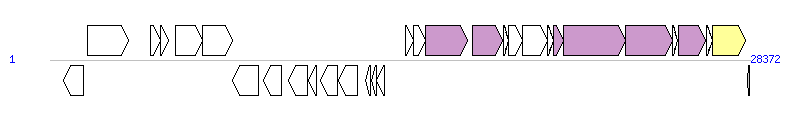 |
| Complete gene list of ICESt3 from AJ586568 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | - | 565..1356 [-], 792 | hypothetical protein | |
| 2 | - | 1520..3192 [+], 1673 | - | |
| 3 | - | 4092..4466 [+], 375 | hypothetical protein | |
| 4 | - | 4483..4785 [+], 303 | hypothetical protein | |
| 5 | - | 5079..6176 [+], 1098 | putative cytosine-specific methyltransferase | |
| 6 | - | 6163..7389 [+], 1227 | putative cytosine-specific methyltransferase | |
| 7 | - | 7414..8457 [-], 1044 | hypothetical protein | |
| 8 | - | 8650..9390 [-], 741 | hypothetical protein | |
| 9 | - | 9659..10447 [-], 789 | - | |
| 10 | - | 10478..10792 [-], 315 | - | |
| 11 | arp1 | 10981..11667 [-], 687 | putative transcriptional regulator | |
| 12 | - | 11676..12479 [-], 804 | putative transcriptional regulator | |
| 13 | - | 12795..12989 [-], 195 | putative transmembrane protein | |
| 14 | - | 13029..13217 [-], 189 | putative transcriptional regulator | |
| 15 | arp2 | 13223..13564 [-], 342 | putative transcriptional regulator | |
| 16 | - | 14427..14723 [+], 297 | putative transfer protein | OrfM; conjugative transfer; [PMID: 15073287] |
| 17 | - | 14745..15206 [+], 462 | putative transfer protein | OrfL; conjugative transfer; [PMID: 15073287] |
| 18 | - | 15220..16908 [+], 1689 | putative transfer protein | YdcQ, T4SS component |
| 19 | - | 17116..18348 [+], 1233 | putative transfer protein | Relaxase, MOBT Family |
| 20 | - | 18363..18593 [+], 231 | hypothetical protein | |
| 21 | - | 18597..19154 [+], 558 | putative transfer protein | OrfH; conjugative transfer; [PMID: 15073287] |
| 22 | - | 19166..20161 [+], 996 | putative transfer protein | OrfG; conjugative transfer; [PMID: 15073287] |
| 23 | - | 20171..20395 [+], 225 | putative transfer protein | OrfF; conjugative transfer; [PMID: 15073287] |
| 24 | - | 20398..20808 [+], 411 | putative transfer protein | YddD, T4SS component |
| 25 | - | 20822..23326 [+], 2505 | putative transfer protein | YddE, T4SS component |
| 26 | - | 23338..25218 [+], 1881 | putative transfer protein | YddG, T4SS component |
| 27 | - | 25220..25444 [+], 225 | hypothetical protein | |
| 28 | - | 25470..26582 [+], 1113 | putative transfer protein | Orf13_p, T4SS component |
| 29 | xis | 26596..26844 [+], 249 | excisionase | Xis; excisionase; [PMID: 15073287] |
| 30 | int | 26844..28190 [+], 1347 | tyrosine integrase | Integrase |
| 31 | fda | 28274..28369 [-], 96 | putative fructose-1,6-diphosphate aldolase | |