
| 44 | |
| SXT(MO10) | |
| ICEO_0000017 | |
| Vibrio cholerae O139 MO10 | |
| 99483 | |
| 47.05 | |
| prfC | |
| Resistance to the antibiotics sulfamethoxazole, trimethoprim, chloramphenicol, and streptomycin; Toxin-Antitoxin System | |
| Vibrio cholerae; Escherichia coli; Salmonella enterica serovar Typhimurium | |
| AY055428 (complete ICE sequence in this GenBank file) | |
| - | |
| 1..99483 | |
| coordinates: 3692..3798; oriTDB id: 200056 TATCGAGACGCCAAACAGTGATTGTTACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGA TAGTGGTTTTGGCTTTTGGGGTTAATTGGATGGGGAA | |
| coordinates: 45176..47326; Gene: traI; Family: MOBH |
The Interaction Network among ICE/IME/CIME | ||||||
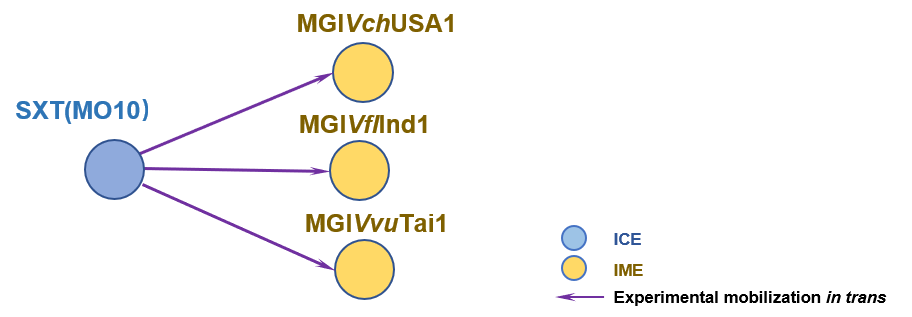 | ||||||
| Detailed Informatioin of the Interaction Network | ||||||
| # | ICE | Inter_Ele | Methods | Donors | Recipients | Exper_Ref |
| 1 | SXT(MO10) | MGIVchUSA1 [IME] | | E. coli CAG18420 | E. coli VB112 | 20807202 |
| 2 | SXT(MO10) | MGIVflInd1 [IME] | | E. coli AD64 | E. coli CAG18439 | 20807202 |
| 3 | SXT(MO10) | MGIVvuTai1 [IME] | | E. coli CAG18420 | E. coli VB112 | 20807202 |
The graph information of SXT(MO10) components from AY055428 | |||||
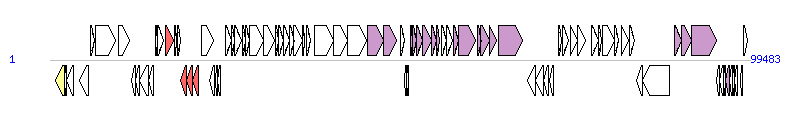 | |||||
| Complete gene list of SXT(MO10) from AY055428 | |||||
| # | Gene | Coordinates [+/-], size (bp) | Product (GenBank annotation) | *Reannotation | |
| 1 | int | 833..2074 [-], 1242 | integrase | Integrase | |
| 2 | s002 (srpR) | 2076..2345 [-], 270 | unknown | SrpR; an active partitioning system; [PMID: 26061412] | |
| 3 | s003 (srpM) | 2348..3322 [-], 975 | unknown | SrpM; an active partitioning system; [PMID: 26061412] | |
| 4 | rumB' | 4241..5413 [-], 1173 | UV repair DNA polymerase | ||
| 5 | tnp | 5697..6290 [+], 594 | putative transposase | ||
| 6 | tnpA | 6404..9304 [+], 2901 | putative transposase | ||
| 7 | tnpB | 9799..11292 [+], 1494 | putative transposase | ||
| 8 | dhfR18 | 11653..12207 [-], 555 | dhfR18 | ||
| 9 | s009 | 12221..12766 [-], 546 | unknown | ||
| 10 | s010 | 12766..13932 [-], 1167 | unknown | ||
| 11 | dcd | 13947..14675 [-], 729 | deoxycytidine triphosphate deaminase | ||
| 12 | 'tnpB | 14935..15267 [+], 333 | putative transposase | ||
| 13 | s013 | 15298..16182 [+], 885 | unknown | ||
| 14 | floR | 16399..17613 [+], 1215 | florfenicol exporter | AR | |
| 15 | s015 | 17641..17946 [+], 306 | unknown | ||
| 16 | tnpB' | 18058..18597 [+], 540 | putative transposase | ||
| 17 | strB | 18569..19405 [-], 837 | streptomycin phosphotransferase | AR | |
| 18 | strA | 19406..20208 [-], 803 | streptomycin phosphotransferase | AR | |
| 19 | sulII | 20269..21084 [-], 816 | dihydropteroate synthase type II | AR | |
| 20 | tnpA' | 21555..23168 [+], 1614 | putative transposase | ||
| 21 | s021 | 22636..23358 [-], 723 | unknown | ||
| 22 | 'rumB | 23511..23831 [-], 321 | UV repair DNA polymerase | ||
| 23 | rumA | 23788..24237 [-], 450 | UV repair | ||
| 24 | s024 | 24876..25784 [+], 909 | unknown | ||
| 25 | s025 | 25858..26172 [+], 315 | unknown | ||
| 26 | s026 | 26241..27167 [+], 927 | unknown | ||
| 27 | s027 | 27395..27778 [+], 384 | unknown | ||
| 28 | s028 | 27766..28359 [+], 594 | unknown | ||
| 29 | s029 | 28378..30273 [+], 1896 | unknown | ||
| 30 | s030 | 30325..31923 [+], 1599 | unknown | ||
| 31 | s031 | 32037..32519 [+], 483 | unknown | ||
| 32 | s032 | 32488..33240 [+], 753 | unknown | ||
| 33 | s033 | 33237..33995 [+], 759 | unknown | ||
| 34 | s034 | 33999..34679 [+], 681 | unknown | ||
| 35 | s035 | 34672..35871 [+], 1200 | unknown | ||
| 36 | s036 | 35852..36232 [+], 381 | unknown | ||
| 37 | s037 | 36399..37013 [+], 615 | unknown | ||
| 38 | s038 | 37601..40234 [+], 2634 | unknown | ||
| 39 | s039 | 40251..42326 [+], 2076 | unknown | ||
| 40 | s040 | 42331..45021 [+], 2691 | unknown | ||
| 41 | traI | 45176..47326 [+], 2151 | conjugative relaxase | Relaxase, MOBH Family | |
| 42 | traD | 47396..49195 [+], 1800 | conjugative coupling factor | TraD_F, T4SS component | |
| 43 | s043 | 49753..50388 [+], 636 | putative conjugation coupling factor | ||
| 44 | s044 | 50414..50692 [-], 279 | unknown | antitoxin, type II toxin-antitoxin system; [PMID: 17158670] | |
| 45 | s045 | 50689..51012 [-], 324 | unknown | toxin, type II toxin-antitoxin system; [PMID: 17158670] | |
| 46 | traL | 51187..51474 [+], 288 | sex pilus assembly | TraL_F, T4SS component | |
| 47 | traE | 51471..52097 [+], 627 | sex pilus assembly | TraE_F, T4SS component | |
| 48 | s048 (traK) | 52078..52977 [+], 900 | unknown | TraK_F, T4SS component | |
| 49 | traB | 52980..54269 [+], 1290 | sex pilus assembly | TraB_F, T4SS component | |
| 50 | traV | 54266..54916 [+], 651 | sex pilus assembly | TraV_F, T4SS component | |
| 51 | traA | 54913..55299 [+], 387 | putative pilin subunit | TraA_F, T4SS component | |
| 52 | s052 (mosA) | 55664..56311 [+], 648 | unknown | MosA; antitoxin, type II toxin-antitoxin system; [PMID: 19325886] | |
| 53 | s053 (mosT) | 56292..57242 [+], 951 | unknown | MosT; toxin, type II toxin-antitoxin system; [PMID: 19325886] | |
| 54 | s054 (dsbC) | 57374..58066 [+], 693 | putative disulfide bond isomerase | TrbB_I, T4SS component | |
| 55 | traC | 58066..60465 [+], 2400 | sex pilus assembly | TraC_F, T4SS component | |
| 56 | trsF | 60789..61301 [+], 513 | conjugation signal peptidase | TraF, T4SS component | |
| 57 | traW | 61216..62436 [+], 1221 | sex pilus assembly | TraW_F, T4SS component | |
| 58 | traU | 62420..63448 [+], 1029 | sex pilus assembly | TraU_F, T4SS component | |
| 59 | traN | 63683..67147 [+], 3465 | mating pair stabilization | TraN_F, T4SS component | |
| 60 | s060 | 67924..69006 [-], 1083 | unknown | ||
| 61 | s061 | 69006..70070 [-], 1065 | unknown | ||
| 62 | s062 | 70183..70899 [-], 717 | unknown | ||
| 63 | s063 | 71008..71610 [-], 603 | unknown | ||
| 64 | ssb | 72318..72737 [+], 420 | single-stranded DNA binding protein | ||
| 65 | s065 (bet) | 72817..73635 [+], 819 | putative DNA recombination protein | Bet; a &lamda; Red-like homologous recombination system; [PMID: 20019796] | |
| 66 | s066 (exo) | 73921..74937 [+], 1017 | unknown | Exo; a &lamda; Red-like homologous recombination system; [PMID: 20019796] | |
| 67 | s067 | 75030..76106 [+], 1077 | unknown | ||
| 68 | s068 | 76975..77928 [+], 954 | unknown | ||
| 69 | s069 | 77990..78430 [+], 441 | unknown | ||
| 70 | s070 | 78500..80155 [+], 1656 | unknown | ||
| 71 | s071 (radC) | 80237..80734 [+], 498 | putative DNA repair radC | RadC; [PMID: 29654185] | |
| 72 | s072 | 81243..82244 [+], 1002 | unknown | ||
| 73 | s073 | 82335..83042 [+], 708 | unknown | ||
| 74 | s074 (dgcL) | 83304..84233 [-], 930 | putative response regulator | DgcL; [PMID: 19888998] | |
| 75 | - | 84239..88045 [-], 3807 | putative histidine kinase | ||
| 76 | traF | 88747..89724 [+], 978 | sex pilus assembly | TraF_F, T4SS component | |
| 77 | traH | 89727..91115 [+], 1389 | sex pilus assembly | TraH_F, T4SS component | |
| 78 | traG | 91119..94688 [+], 3570 | sex pilus assembly | TraG_F, T4SS component | |
| 79 | s079 (eexS) | 94721..95173 [-], 453 | unknown | EexS; entry exclusion factor; [PMID: 15935784; 29654185] | |
| 80 | setC | 95207..95740 [-], 534 | transcriptional activator | ||
| 81 | setD | 95737..96036 [-], 300 | transcriptional activator | ||
| 82 | s082 | 96033..96581 [-], 549 | unknown | Orf169_F, T4SS component | |
| 83 | s083 | 96568..96936 [-], 369 | unknown | ||
| 84 | s084 | 96926..97231 [-], 306 | unknown | ||
| 85 | s085 | 97218..97739 [-], 522 | unknown | ||
| 86 | s086 (croS) | 98144..98395 [-], 252 | unknown | CroS; transcriptional repressors; [PMID: 29654185] | |
| 87 | setR | 98513..99160 [+], 648 | transcriptional repressor | ||
|
(1) Daccord A; Mursell M; Poulin-Laprade D; Burrus V (2012). Dynamics of the SetCD-Regulated Integration and Excision of Genomic Islands Mobilized by Integrating Conjugative Elements of the SXT/R391 Family. J Bacteriol. 194(21):5794-802. [PubMed:22923590] |
|
(2) Pande K; Mendiratta DK; Vijayashri D; Thamke DC; Narang P (2012). SXT constin among Vibrio cholerae isolates from a tertiary care hospital. Indian J Med Res. 135:346-50. [PubMed:22561621] |
|
(3) Spagnoletti M; Ceccarelli D; Colombo MM (2012). Rapid detection by multiplex PCR of Genomic Islands, prophages and Integrative Conjugative Elements in V. cholerae 7th pandemic variants. J Microbiol Methods. 88(1):98-102. [PubMed:22062086] |
|
(4) Chen WY; Ho JW; Huang JD; Watt RM (2011). Functional characterization of an alkaline exonuclease and single strand annealing protein from the SXT genetic element of Vibrio cholerae. BMC Mol Biol. 12(1):16. [PubMed:21501469] |
|
(5) Wozniak RA; Fouts DE; Spagnoletti M; Colombo MM; Ceccarelli D; Garriss G; Dery C; Burrus V; Waldor MK (2009). Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet. 5(12):e1000786. [PubMed:20041216] |
|
(6) Bordeleau E; Brouillette E; Robichaud N; Burrus V (2010). Beyond antibiotic resistance: integrating conjugative elements of the SXT/R391 family that encode novel diguanylate cyclases participate to c-di-GMP signalling in Vibrio cholerae. Environ Microbiol. 12(2):510-23. [PubMed:19888998] |
|
(7) Wozniak RA; Waldor MK (2009). A toxin-antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genet. 5(3):e1000439. [PubMed:19325886] |
|
(8) Ceccarelli D; Daccord A; Rene M; Burrus V (2008). Identification of the origin of transfer (oriT) and a new gene required for mobilization of the SXT/R391 family of integrating conjugative elements. J Bacteriol. 190(15):5328-38. [PubMed:18539733] |
|
(9) Marrero J; Waldor MK (2007). The SXT/R391 family of integrative conjugative elements is composed of two exclusion groups. J Bacteriol. 189(8):3302-5. [PubMed:17307849] |
|
(10) McLeod SM; Burrus V; Waldor MK (2006). Requirement for Vibrio cholerae integration host factor in conjugative DNA transfer. J Bacteriol. 188(16):5704-11. [PubMed:16885438] |
|
(11) Marrero J; Waldor MK (2005). Interactions between inner membrane proteins in donor and recipient cells limit conjugal DNA transfer. Dev Cell. 8(6):963-70. [PubMed:15935784] |
|
(12) Beaber JW; Waldor MK (2004). Identification of operators and promoters that control SXT conjugative transfer. J Bacteriol. 186(17):5945-9. [PubMed:15317801] |
|
(13) Burrus V; Waldor MK (2004). Formation of SXT tandem arrays and SXT-R391 hybrids. J Bacteriol. 186(9):2636-45. [PubMed:15090504] |
|
(14) Boltner D; Osborn AM (2004). Structural comparison of the integrative and conjugative elements R391, pMERPH, R997, and SXT. Plasmid. 51(1):12-23. [PubMed:14711525] |
|
(15) Beaber JW; Hochhut B; Waldor MK (2004). SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature. 427(6969):72-4. [PubMed:14688795] |
|
(16) Burrus V; Waldor MK (2003). Control of SXT integration and excision. J Bacteriol. 185(17):5045-54. [PubMed:12923077] |
|
(17) Thungapathra M; Amita; Sinha KK; Chaudhuri SR; Garg P; Ramamurthy T; Nair GB; Ghosh A (2002). Occurrence of antibiotic resistance gene cassettes aac(6')-Ib, dfrA5, dfrA12, and ereA2 in class I integrons in non-O1, non-O139 Vibrio cholerae strains in India. Antimicrob Agents Chemother. 46(9):2948-55. [PubMed:12183252] |
|
(18) Beaber JW; Hochhut B; Waldor MK (2002). Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J Bacteriol. 184(15):4259-69. [PubMed:12107144] |
|
(19) Hochhut B; Lotfi Y; Mazel D; Faruque SM; Woodgate R; Waldor MK (2001). Molecular analysis of antibiotic resistance gene clusters in vibrio cholerae O139 and O1 SXT constins. Antimicrob Agents Chemother. 45(11):2991-3000. [PubMed:11600347] |
|
(20) Hochhut B; Beaber JW; Woodgate R; Waldor MK (2001). Formation of chromosomal tandem arrays of the SXT element and R391, two conjugative chromosomally integrating elements that share an attachment site. J Bacteriol. 183(4):1124-32. [PubMed:11157923] |
|
(21) Hochhut B; Marrero J; Waldor MK (2000). Mobilization of plasmids and chromosomal DNA mediated by the SXT element, a constin found in Vibrio cholerae O139. J Bacteriol. 182(7):2043-7. [PubMed:10715015] |
|
(22) Hochhut B; Waldor MK (1999). Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol Microbiol. 32(1):99-110. [PubMed:10216863] |
|
(23) Waldor MK; Tschape H; Mekalanos JJ (1996). A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J Bacteriol. 178(14):4157-65. [PubMed:8763944] |