Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 100253 |
| Name | oriT_pCF10 |
| Organism | Enterococcus faecalis |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NC_006827 (32815..32864 [+], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..16, 17..26 (GCAACATGCT..AGCATGTTGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
GCTTGCAAAA |
| Note |
oriT sequence
Download Length: 50 nt
>oriT_pCF10
AATATCGCAACATGCTAGCATGTTGCTCCGCTTGCAAAAAGAAAGCCTAC
AATATCGCAACATGCTAGCATGTTGCTCCGCTTGCAAAAAGAAAGCCTAC
Visualization of oriT structure (The oriT was characterized experimentally)
oriT secondary structure
Predicted by RNAfold.
Download structure file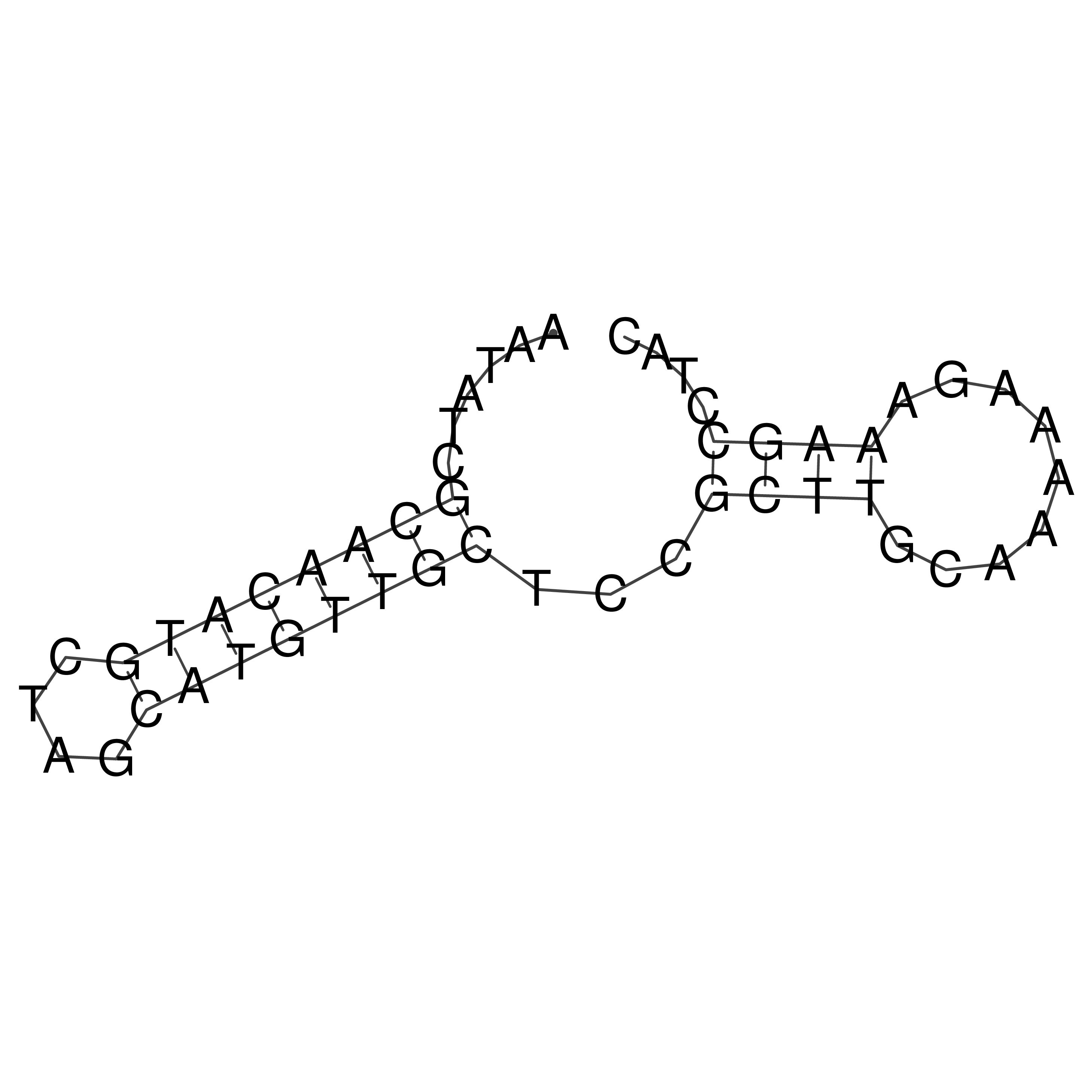
Reference
[1] Saima Rehman et al. (2019) Enterococcal PcfF Is a Ribbon-Helix-Helix Protein That Recruits the Relaxase PcfG Through Binding and Bending of the oriT Sequence. Frontiers in microbiology. 1.081944444. [PMID:31134011]
Relaxosome
This oriT is a component of a relaxosome.
| Relaxosome name | RelaxosomepCF10 |
| oriT | oriT_pCF10 |
| Relaxase | PcfG_pCF10 |
| Auxiliary protein | PcfF_pCF10 |
Reference
[1] Saima Rehman et al. (2019) Enterococcal PcfF Is a Ribbon-Helix-Helix Protein That Recruits the Relaxase PcfG Through Binding and Bending of the oriT Sequence. Frontiers in microbiology. 1.081944444. [PMID:31134011]
Relaxase
| ID | 324 | GenBank | YP_195793 |
| Name | PcfG_pCF10 |
UniProt ID | Q7X5P3 |
| Length | 561 a.a. | PDB ID | |
| Note | putative relaxase | ||
Relaxase protein sequence
Download Length: 561 a.a. Molecular weight: 65380.01 Da Isoelectric Point: 6.2230
>YP_195793.1 PcfG (plasmid) [Enterococcus faecalis]
MVYTKHFVIHTFDKLNNACSYIENAEKTEVTNDNPSEHLEHLFQYIVNDDKTYMKKLVSGHGIVDPTNPY
EEFKLTKLQAAIQRKIGYTFDPKSERLLPPTLTELEKGNAVLAHHLIQSFSPEDDLTPEKIHEIGYNTVM
ELTGGKYEFVIATHVDKEHLHNHIIFSSTNLKTGKAFRWQKGTKRVFEQISDKIAAKEGAKIIEKSPKQT
HTKYTKWQTESIYKQKIKSRLDFLLEQSSSINDFLEKAEALNLSADFSNKWTTYRLLDEPQIKNTRSRSL
SKSDPTRYNYEKIIERLKENKNVLTVEECVKRYEEKNEQEKNNFDYQFTIEPWQLSHKTERGYYINIDYG
YGNSGKLFVGGYKVDPLDNGNYNIYIKRNEYFYFMSDKDATKSKYLTGASLIKQLRLYNGQTPLKREPVM
QTIDELVSAINFLAANEIEDTRQLKLLEEKLEVAFLEAEKTLETLDEKMLELHQLSNLLLENELQGDPEL
IQKKLKTLLPDATLAEFSYEDVRGEIEAIKTSQTLLESKLERTRNEINQLHEIQAVQEKEPEKQTDIKPK
L
MVYTKHFVIHTFDKLNNACSYIENAEKTEVTNDNPSEHLEHLFQYIVNDDKTYMKKLVSGHGIVDPTNPY
EEFKLTKLQAAIQRKIGYTFDPKSERLLPPTLTELEKGNAVLAHHLIQSFSPEDDLTPEKIHEIGYNTVM
ELTGGKYEFVIATHVDKEHLHNHIIFSSTNLKTGKAFRWQKGTKRVFEQISDKIAAKEGAKIIEKSPKQT
HTKYTKWQTESIYKQKIKSRLDFLLEQSSSINDFLEKAEALNLSADFSNKWTTYRLLDEPQIKNTRSRSL
SKSDPTRYNYEKIIERLKENKNVLTVEECVKRYEEKNEQEKNNFDYQFTIEPWQLSHKTERGYYINIDYG
YGNSGKLFVGGYKVDPLDNGNYNIYIKRNEYFYFMSDKDATKSKYLTGASLIKQLRLYNGQTPLKREPVM
QTIDELVSAINFLAANEIEDTRQLKLLEEKLEVAFLEAEKTLETLDEKMLELHQLSNLLLENELQGDPEL
IQKKLKTLLPDATLAEFSYEDVRGEIEAIKTSQTLLESKLERTRNEINQLHEIQAVQEKEPEKQTDIKPK
L
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q7X5P3 |
Host bacterium
| ID | 1462 | GenBank | NC_006827 |
| Plasmid name | pCF10 | Incompatibility group | - |
| Plasmid size | 67673 bp | Coordinate of oriT [Strand] | 32815..32864 [+] |
| Host baterium | Enterococcus faecalis |
Cargo genes
| Drug resistance gene | resistance to doxycycline, tetracycline and minocycline |
| Virulence gene | prgB/asc10 |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21, AcrIIA9 |