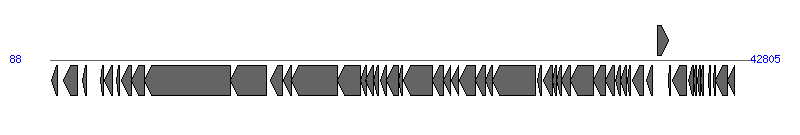The information of T4SS components from NC_010488 |
 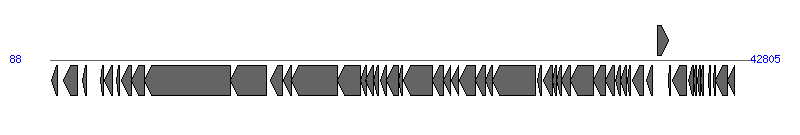 |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | EcSMS35_A0001 | 154..540 [-], 387 | 170650844 | replication initiation protein repA4 | | |
| 2 | EcSMS35_A0002 (repA1) | 904..1773 [-], 870 | 170650780 | replication protein | | |
| 3 | EcSMS35_A0003 (repA2) | 2054..2314 [-], 261 | 170650796 | replication protein | | |
| 4 | EcSMS35_A0005 | 3164..3373 [-], 210 | 170650792 | hypothetical protein | | |
| 5 | EcSMS35_A0006 | 3419..3880 [-], 462 | 170650855 | nuclease homolog | | |
| 6 | EcSMS35_A0007 | 4123..4335 [-], 213 | 170650817 | hypothetical protein | | |
| 7 | EcSMS35_A0008 (finO) | 4473..5084 [-], 612 | 170650904 | conjugal transfer fertility inhibition protein FinO | | |
| 8 | EcSMS35_A0009 (traX) | 5088..5834 [-], 747 | 170650820 | conjugal transfer pilus acetylation protein TraX | TraX_F | |
| 9 | EcSMS35_A0010 (traI) | 5854..11124 [-], 5271 | 170650784 | conjugal transfer nickase/helicase TraI | TraI_F | |
| 10 | EcSMS35_A0011 (traD) | 11124..13295 [-], 2172 | 170650889 | conjugal transfer protein TraD | TraD_F | |
| 11 | EcSMS35_A0012 (traT) | 13548..14279 [-], 732 | 170650825 | conjugal transfer surface exclusion protein TraT | | |
| 12 | EcSMS35_A0013 | 14311..14808 [-], 498 | 170650886 | putative type IV secretion-like conjugative transfer system protein TraS | | |
| 13 | EcSMS35_A0014 (traG) | 14829..17651 [-], 2823 | 170650902 | conjugal transfer mating pair stabilization protein TraG | TraG_F | |
| 14 | EcSMS35_A0015 (traH) | 17648..19024 [-], 1377 | 170650800 | conjugal transfer pilus assembly protein TraH | TraH_F | |
| 15 | EcSMS35_A0016 (trbJ) | 19021..19383 [-], 363 | 170650880 | conjugal transfer protein TrbJ | | |
| 16 | EcSMS35_A0017 (trbB) | 19313..19858 [-], 546 | 170650781 | conjugal transfer protein TrbB | | |
| 17 | EcSMS35_A0018 (traQ) | 19845..20129 [-], 285 | 170650897 | conjugal transfer pilin chaperone TraQ | TraQ_F | |
| 18 | EcSMS35_A0019 (trbA) | 20248..20595 [-], 348 | 170650822 | conjugal transfer protein TrbA | | |
| 19 | EcSMS35_A0020 (traF) | 20611..21354 [-], 744 | 170650828 | conjugal pilus assembly protein TraF | TraF_F | |
| 20 | EcSMS35_A0021 (trbE) | 21347..21604 [-], 258 | 170650840 | conjugal transfer protein TrbE | | |
| 21 | EcSMS35_A0022 (traN) | 21631..23439 [-], 1809 | 170650771 | conjugal transfer mating pair stabilization protein TraN | TraN_F | |
| 22 | EcSMS35_A0023 (trbC) | 23436..24074 [-], 639 | 170650809 | conjugal transfer pilus assembly protein TrbC | TrbC_F | |
| 23 | EcSMS35_A0024 | 24102..24554 [-], 453 | 170650862 | hypothetical protein | | |
| 24 | EcSMS35_A0025 | 24584..25045 [-], 462 | 170650772 | hypothetical protein | | |
| 25 | EcSMS35_A0026 (traU) | 25071..26063 [-], 993 | 170650911 | conjugal transfer pilus assembly protein TraU | TraU_F | |
| 26 | EcSMS35_A0027 (traW) | 26060..26692 [-], 633 | 170650832 | conjugal transfer pilus assembly protein TraW | TraW_F | |
| 27 | EcSMS35_A0028 (trbI) | 26689..27075 [-], 387 | 170650810 | conjugal transfer protein TrbI | | |
| 28 | EcSMS35_A0029 (traC) | 27072..29699 [-], 2628 | 170650779 | conjugal transfer ATP-binding protein TraC | TraC_F | |
| 29 | EcSMS35_A0030 (traR) | 29859..30080 [-], 222 | 170650873 | conjugal transfer protein TraR | | |
| 30 | EcSMS35_A0031 (traV) | 30215..30730 [-], 516 | 170650794 | conjugal transfer protein TraV | TraV_F | |
| 31 | EcSMS35_A0032 | 30727..30978 [-], 252 | 170650806 | conjugal transfer protein TrbG | | |
| 32 | EcSMS35_A0033 | 30975..31292 [-], 318 | 170650884 | conjugal transfer protein TrbD | | |
| 33 | EcSMS35_A0034 (traP) | 31289..31861 [-], 573 | 170650891 | conjugal transfer protein TraP | | |
| 34 | EcSMS35_A0035 (traB) | 31851..33278 [-], 1428 | 170650812 | conjugal transfer pilus assembly protein TraB | TraB_F | |
| 35 | EcSMS35_A0036 (traK) | 33278..34006 [-], 729 | 170650906 | conjugal transfer protein TraK | TraK_F | |
| 36 | EcSMS35_A0037 (traE) | 33993..34559 [-], 567 | 170650797 | conjugal transfer pilus assembly protein TraE | TraE_F | |
| 37 | EcSMS35_A0038 (traL) | 34581..34892 [-], 312 | 170650856 | conjugal transfer pilus assembly protein TraL | TraL_F | |
| 38 | EcSMS35_A0039 (traA) | 34907..35266 [-], 360 | 170650824 | conjugal transfer pilin subunit TraA | TraA_F | |
| 39 | EcSMS35_A0040 (traY) | 35300..35527 [-], 228 | 170650869 | conjugal transfer protein TraY | TraY_F | |
| 40 | EcSMS35_A0041 (traJ) | 35621..36307 [-], 687 | 170650783 | type IV secretion-like conjugative transfer system regulator TraJ | | |
| 41 | EcSMS35_A0042 (traM) | 36498..36881 [-], 384 | 170650851 | conjugal transfer protein TraM | TraM_F |  |
| 42 | EcSMS35_A0043 | 37158..37805 [+], 648 | 170650864 | lytic transglycosylase | Orf169_F | |
| 43 | EcSMS35_A0044 | 37831..37965 [-], 135 | 170650793 | hypothetical protein | | |
| 44 | EcSMS35_A0045 | 38102..38923 [-], 822 | 170650799 | hypothetical protein | | |
| 45 | EcSMS35_A0046 | 39046..39333 [-], 288 | 170650769 | hypothetical protein | | |
| 46 | EcSMS35_A0047 | 39358..39519 [-], 162 | 170650803 | hypothetical protein | | |
| 47 | EcSMS35_A0048 | 39600..39812 [-], 213 | 170650770 | hypothetical protein | | |
| 48 | EcSMS35_A0049 | 39805..39957 [-], 153 | 170650839 | hypothetical protein | | |
| 49 | EcSMS35_A0050 (hok) | 40255..40413 [-], 159 | 170650845 | small toxic polypeptide | | |
| 50 | EcSMS35_A0051 | 40556..40693 [-], 138 | 170650801 | hypothetical protein | | |
| 51 | EcSMS35_A0052 (psiA) | 40693..41412 [-], 720 | 170650870 | plasmid SOS inhibition protein A | | |
| 52 | EcSMS35_A0053 (psiB) | 41409..41843 [-], 435 | 170650814 | plasmid SOS inhibition protein B | | |
| |