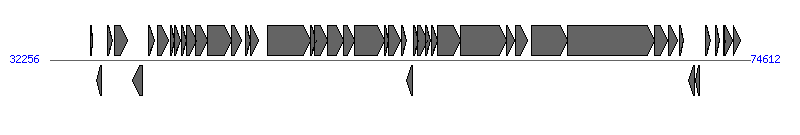The information of T4SS components from NC_010558 |
 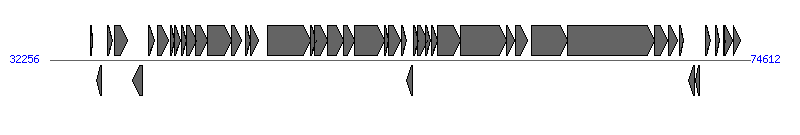 |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | IPF_336 | 34676..34834 [+], 159 | 187736776 | small toxic polypeptide | | |
| 2 | IPF_334 (YubN) | 35072..35389 [-], 318 | 187736777 | YubN | | |
| 3 | IPF_331 (YubO) | 35733..36020 [+], 288 | 187736778 | YubO | | |
| 4 | IPF_329 (YubP) | 36139..36960 [+], 822 | 187736779 | YubP | | |
| 5 | IPF_328 | 37256..37846 [-], 591 | 187736780 | lytic transglycosylase | Orf169_F | |
| 6 | IPF_327 (TraM) | 38198..38581 [+], 384 | 187736781 | conjugal transfer protein TraM | TraM_F |  |
| 7 | IPF_326 (TraJ) | 38772..39419 [+], 648 | 187736782 | conjugal transfer transcriptional regulator TraJ | | |
| 8 | IPF_396 (TraY) | 39555..39770 [+], 216 | 187736783 | conjugal transfer protein TraY | TraY_F | |
| 9 | IPF_325 (TraA) | 39813..40178 [+], 366 | 187736784 | conjugal transfer pilin subunit TraA | TraA_F | |
| 10 | IPF_323 (TraL) | 40193..40504 [+], 312 | 187736785 | conjugal transfer pilus assembly protein TraL | TraL_F | |
| 11 | IPF_322 (TraE) | 40526..41092 [+], 567 | 187736786 | conjugal transfer pilus assembly protein TraE | TraE_F | |
| 12 | IPF_321 (TraK) | 41079..41807 [+], 729 | 187736787 | conjugal transfer protein TraK | TraK_F | |
| 13 | IPF_318 (TraB) | 41807..43234 [+], 1428 | 187736788 | conjugal transfer pilus assembly protein TraB | TraB_F | |
| 14 | IPF_315 (TraP) | 43224..43814 [+], 591 | 187736789 | conjugal transfer protein TraP | | |
| 15 | IPF_314 (TrbG) | 44114..44365 [+], 252 | 187736790 | conjugal transfer protein TrbG | | |
| 16 | IPF_312 (TraV) | 44362..44877 [+], 516 | 187736791 | conjugal transfer protein TraV | TraV_F | |
| 17 | IPF_309 (TraC) | 45393..48020 [+], 2628 | 187736792 | conjugal transfer ATP-binding protein TraC | TraC_F | |
| 18 | IPF_306a (TrbI) | 48017..48403 [+], 387 | 187736793 | conjugal transfer protein TrbI | | |
| 19 | IPF_304 (TraW) | 48286..49032 [+], 747 | 187736794 | conjugal transfer pilus assembly protein TraW | TraW_F | |
| 20 | IPF_303 (TraU) | 49029..50021 [+], 993 | 187736795 | conjugal transfer pilus assembly protein TraU | TraU_F | |
| 21 | IPF_301 (TrbC) | 50027..50668 [+], 642 | 187736796 | conjugal transfer pilus assembly protein TrbC | TrbC_F | |
| 22 | IPF_299 (TraN) | 50665..52473 [+], 1809 | 187736797 | conjugal transfer mating pair stabilization protein TraN | TraN_F | |
| 23 | IPF_397 (TrbE) | 52497..52757 [+], 261 | 187736798 | conjugal transfer protein TrbE | | |
| 24 | IPF_297 (TraF) | 52720..53493 [+], 774 | 187736799 | conjugal pilus assembly protein TraF | TraF_F | |
| 25 | IPF_295 (TrbA) | 53507..53848 [+], 342 | 187736800 | conjugal transfer protein TrbA | | |
| 26 | IPF_294 (ArtA) | 53829..54164 [-], 336 | 187736801 | ArtA | | |
| 27 | IPF_293 (TraQ) | 54245..54529 [+], 285 | 187736802 | conjugal transfer pilin chaperone TraQ | TraQ_F | |
| 28 | IPF_291 (TrbB) | 54516..55070 [+], 555 | 187736803 | conjugal transfer protein TrbB | | |
| 29 | IPF_405 (TrbJ) | 55000..55284 [+], 285 | 187736804 | conjugal transfer protein TrbJ | | |
| 30 | IPF_290 (TrbF) | 55327..55719 [+], 393 | 187736805 | conjugal transfer protein TrbF | | |
| 31 | IPF_289 (TraH) | 55703..57079 [+], 1377 | 187736806 | conjugal transfer pilus assembly protein TraH | TraH_F | |
| 32 | IPF_286 (TraG) | 57076..59901 [+], 2826 | 187736807 | conjugal transfer mating pair stabilization protein TraG | TraG_F | |
| 33 | IPF_284 (TraS) | 59898..60353 [+], 456 | 187736808 | conjugal transfer entry exclusion protein TraS | | |
| 34 | IPF_283 (TraT) | 60422..61153 [+], 732 | 187736809 | conjugal transfer surface exclusion protein TraT | | |
| 35 | IPF_282 (TraD) | 61405..63576 [+], 2172 | 187736810 | conjugal transfer protein TraD | TraD_F | |
| 36 | IPF_279a (TraI) | 63576..68846 [+], 5271 | 187736811 | conjugal transfer nickase/helicase TraI | TraI_F | |
| 37 | IPF_270a (TraX) | 68809..69612 [+], 804 | 187736812 | conjugal transfer pilus acetylation protein TraX | TraX_F | |
| 38 | IPF_268 (FinO) | 69667..70227 [+], 561 | 187736813 | conjugal transfer fertility inhibition protein FinO | | |
| 39 | IPF_267 (YigA) | 70360..70572 [+], 213 | 187736814 | YigA | | |
| 40 | IPF_265 (IS1-insB) | 70873..71259 [-], 387 | 187736815 | IS1-insB | | |
| 41 | IPF_264 (IS1-insA) | 71300..71575 [-], 276 | 187736816 | IS1-insA | | |
| 42 | IPF_386 (SrnB) | 71940..72251 [+], 312 | 187736817 | SrnB | | |
| 43 | IPF_384 (RepA2) | 72535..72792 [+], 258 | 187736818 | replication protein | | |
| 44 | IPF_387 (RepA6) | 73028..73102 [+], 75 | 187736819 | leader peptide RepL | | |
| 45 | IPF_260a (RepA1) | 73095..73547 [+], 453 | 187736820 | RepA1 | | |
| 46 | IPF_258 | 73596..74051 [+], 456 | 187736821 | hypothetical protein | | |
| |