
| 8 | |
| Plasmid F | |
| plasmid F [Browse all T4SS(s) in this replicon] | |
| NC_002483 | |
| 65556..98709 | |
| Tra_F | |
| Conjugation | |
| Type IVA; Type F | |
T4SS components |
| Component | TraA_F | TraB_F | TraC_F | TraD_F | TraE_F | TraF_F | TraG_F | TraH_F | TraI_F |
| Number | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Component | TraK_F | TraL_F | TraM_F | TraN_F | TraQ_F | TraU_F | TraV_F | TraW_F | TraX_F |
| Number | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Component | TraY_F | TrbC_F | Orf169_F | ||||||
| Number | 1 | 1 | 1 |
The information of T4SS components from NC_002483 | ||||||
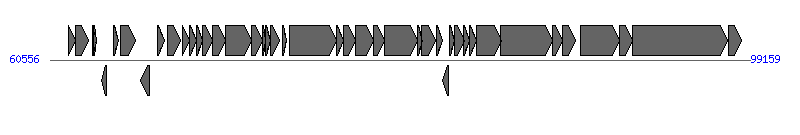 | ||||||
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | D616_p97097 (psiB) | 61551..61985 [+], 435 | 9507776 | hypothetical protein | ||
| 2 | D616_p97098 (psiA) | 61982..62701 [+], 720 | 9507777 | hypothetical protein | ||
| 3 | D616_p97102 (flmC) | 62918..63130 [+], 213 | 9507778 | hypothetical protein | ||
| 4 | D616_p97103 (flmA) | 62976..63134 [+], 159 | 9507779 | hypothetical protein | ||
| 5 | D616_p97004 (ygdA) | 63372..63689 [-], 318 | 9507780 | hypothetical protein | ||
| 6 | D616_p97003 (ygeA) | 64032..64319 [+], 288 | 9507781 | hypothetical protein | ||
| 7 | D616_p97002 (ygeB) | 64438..65259 [+], 822 | 9507782 | hypothetical protein | ||
| 8 | D616_p97001 (ygfA) | 65556..66065 [-], 510 | 9507783 | hypothetical protein | Orf169_F | |
| 9 | D616_p97066 (traM) | 66479..66862 [+], 384 | 9507784 | hypothetical protein | TraM_F | |
| 10 | D616_p97072 (traJ) | 67049..67738 [+], 690 | 9507785 | hypothetical protein | ||
| 11 | D616_p97057 (traY) | 67837..68232 [+], 396 | 9507786 | hypothetical protein | TraY_F | |
| 12 | D616_p97080 (traA) | 68265..68630 [+], 366 | 9507787 | prepropilin | TraA_F | |
| 13 | D616_p97069 (traL) | 68645..68956 [+], 312 | 9507788 | hypothetical protein | TraL_F | |
| 14 | D616_p97074 (traE) | 68978..69544 [+], 567 | 9507789 | hypothetical protein | TraE_F | |
| 15 | D616_p97071 (traK) | 69531..70259 [+], 729 | 9507790 | hypothetical protein | TraK_F | |
| 16 | D616_p97076 (traB) | 70259..71686 [+], 1428 | 9507791 | hypothetical protein | TraB_F | |
| 17 | D616_p97064 (traP) | 71676..72266 [+], 591 | 9507792 | hypothetical protein | ||
| 18 | D616_p97051 (trbD) | 72253..72450 [+], 198 | 9507793 | hypothetical protein | ||
| 19 | D616_p97049 (trbG) | 72462..72713 [+], 252 | 9507794 | hypothetical protein | ||
| 20 | D616_p97059 (traV) | 72710..73225 [+], 516 | 9507795 | hypothetical protein | TraV_F | |
| 21 | D616_p97062 (traR) | 73360..73581 [+], 222 | 9507796 | hypothetical protein | ||
| 22 | D616_p97075 (traC) | 73741..76368 [+], 2628 | 9507797 | hypothetical protein | TraC_F | |
| 23 | D616_p97048 (trbI) | 76365..76751 [+], 387 | 9507798 | hypothetical protein | ||
| 24 | D616_p97058 (traW) | 76748..77380 [+], 633 | 9507799 | hypothetical protein | TraW_F | |
| 25 | D616_p97061 (traU) | 77377..78369 [+], 993 | 9507800 | hypothetical protein | TraU_F | |
| 26 | D616_p97053 (trbC) | 78378..79016 [+], 639 | 9507801 | hypothetical protein | TrbC_F | |
| 27 | D616_p97065 (traN) | 79013..80821 [+], 1809 | 9507802 | hypothetical protein | TraN_F | |
| 28 | D616_p97050 (trbE) | 80845..81105 [+], 261 | 9507803 | hypothetical protein | ||
| 29 | D616_p97073 (traF) | 81068..81841 [+], 774 | 9507804 | hypothetical protein | TraF_F | |
| 30 | D616_p97055 (trbA) | 81857..82204 [+], 348 | 9507805 | hypothetical protein | ||
| 31 | D616_p97107 (artA) | 82206..82520 [-], 315 | 9507806 | hypothetical protein | ||
| 32 | D616_p97070 (traQ) | 82601..82885 [+], 285 | 9507807 | hypothetical protein | TraQ_F | |
| 33 | D616_p97060 (trbB) | 82872..83417 [+], 546 | 9507808 | hypothetical protein | ||
| 34 | D616_p97052 (trbJ) | 83347..83688 [+], 342 | 9507809 | hypothetical protein | ||
| 35 | D616_p97056 (trbF) | 83675..84055 [+], 381 | 9507810 | hypothetical protein | ||
| 36 | D616_p97078 (traH) | 84052..85428 [+], 1377 | 9507811 | hypothetical protein | TraH_F | |
| 37 | D616_p97079 (traG) | 85425..88241 [+], 2817 | 9507812 | hypothetical protein | TraG_F | |
| 38 | D616_p97068 (traS) | 88274..88795 [+], 522 | 9507813 | surface exclusion protien | ||
| 39 | D616_p97067 (traT) | 88817..89551 [+], 735 | 9507814 | surface exclusion protien | ||
| 40 | D616_p97082 (traD) | 89804..91957 [+], 2154 | 9507815 | hypothetical protein | TraD_F | |
| 41 | D616_p97054 (trbH) | 91957..92676 [+], 720 | 9507816 | hypothetical protein | ||
| 42 | D616_p97077 (traI) | 92673..97943 [+], 5271 | 9507817 | hypothetical protein | TraI_F | |
| 43 | D616_p97063 (traX) | 97963..98709 [+], 747 | 9507818 | hypothetical protein | TraX_F | |
Download FASTA format files |
| Proteins Genes |
| # | Name | Image | Resource | Detail | Reference |
| 1 | TraI | 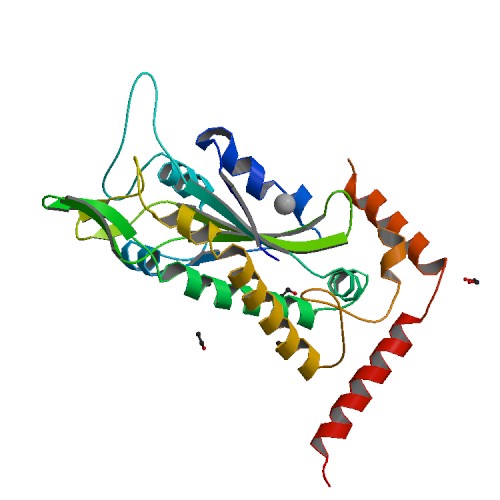 | PDB (1P4D) | F factor TraI Relaxase Domain. | (1) PubMed: 14604527 |
| 2 | TraI (381-569) | 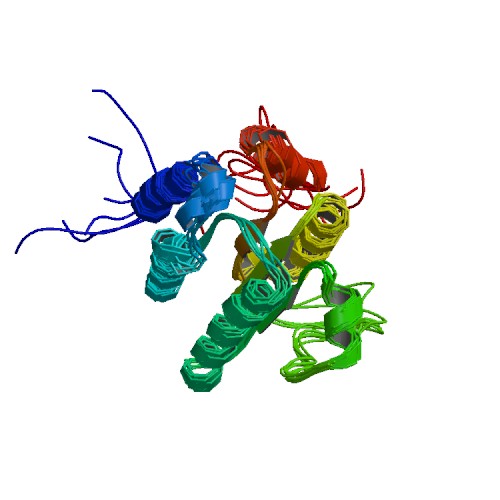 | PDB (2L8B) | TraI (381-569) | (2) PubMed: 22611034 |
| 3 | TraI bound to ssDNA | 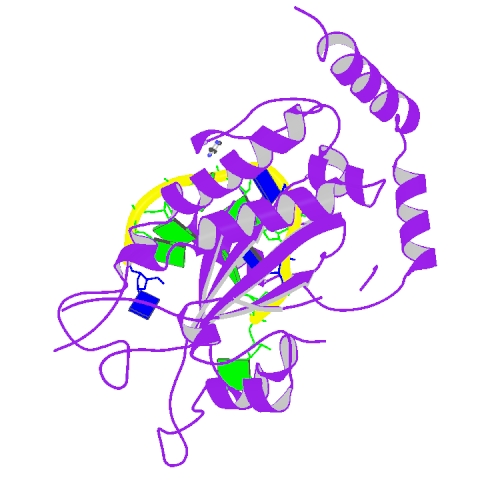 | PDB (2A0I) | F Factor TraI Relaxase Domain bound to F oriT Single-stranded DNA. | (3) PubMed: 16216584 |
| 4 | TraI c-terminal domain | 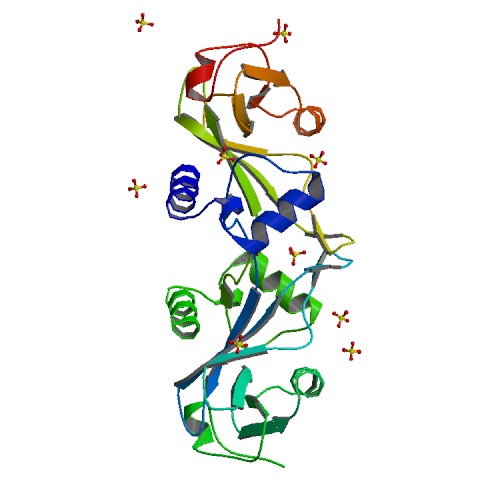 | PDB (3FLD) | Crystal structure of the trai c-terminal domain. | (4) PubMed: 19136009 |
| 5 | TraI with the Scissile Thymidine Base | 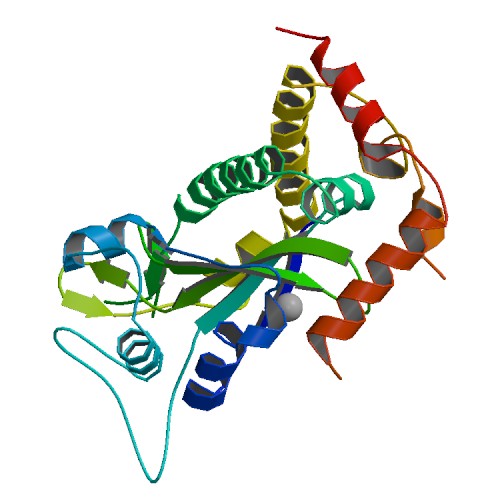 | PDB (2Q7T) | Crystal Structure of the F plasmid TraI Relaxase Domain with the Scissile Thymidine Base. | (5) PubMed: 17630285 |
| 6 | TraI with the Scissile Thymidine Base and Imidodiphosphate | 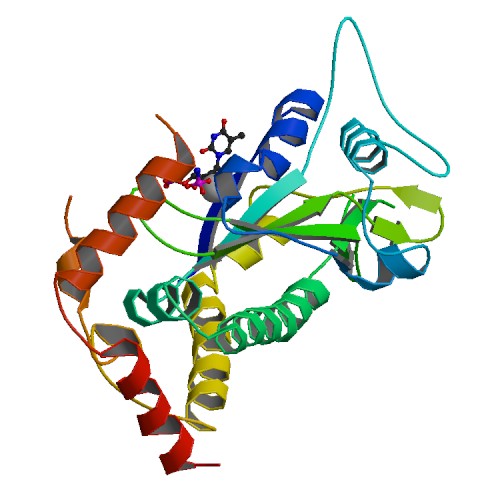 | PDB (2Q7U) | Crystal Structure of the F plasmid TraI Relaxase Domain with the Scissile Thymidine Base and Imidodiphosphate. | (6) PubMed: 17630285 |
| 7 | TraM-traD | 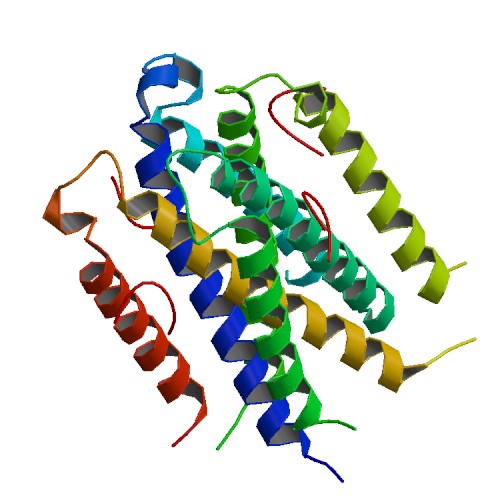 | PDB (3D8A) | Co-crystal structure of TraM-TraD complex. | (7) PubMed: 18717787 |
|
(1) Datta S et al. (2003). Structural insights into single-stranded DNA binding and cleavage by F factor TraI. Structure. 11(11):1369-79. [PudMed:14604527] |
|
(2) Wright NT et al. (2012). Solution structure and small angle scattering analysis of TraI (381-569). Proteins. 80(9):2250-61. [PudMed:22611034] |
|
(3) Larkin C et al. (2005). Inter- and intramolecular determinants of the specificity of single-stranded DNA binding and cleavage by the F factor relaxase. Structure. 13(10):1533-44.. [PudMed:16216584] |
|
(4) Guogas LM et al. (2009). A novel fold in the TraI relaxase-helicase c-terminal domain is essential for conjugative DNA transfer. J Mol Biol. 386(2):554-68. [PudMed:19136009] |
|
(5) Lujan SA et al. (2007). Disrupting antibiotic resistance propagation by inhibiting the conjugative DNA relaxase. Proc Natl Acad Sci U S A. 104(30):12282-7. [PudMed:17630285] |
|
(6) Lujan SA et al. (2007). Disrupting antibiotic resistance propagation by inhibiting the conjugative DNA relaxase. Proc Natl Acad Sci U S A. 104(30):12282-7. [PudMed:17630285] |
|
(7) Lu J et al. (2008). Structural basis of specific TraD-TraM recognition during F plasmid-mediated bacterial conjugation. Mol Microbiol. 70(1):89-99. [PudMed:18717787] |
| (1) Frost LS; Koraimann G (2010). Regulation of bacterial conjugation: balancing opportunity with adversity. Future Microbiol. 5(7):1057-71. [PudMed:20632805] |
|
(2) Dorman CJ (2009). Nucleoid-associated proteins and bacterial physiology. Adv Appl Microbiol. 67:47-64. [PudMed:19245936] |
|
(3) Audette GF; Manchak J; Beatty P; Klimke WA; Frost LS (2007). Entry exclusion in F-like plasmids requires intact TraG in the donor that recognizes its cognate TraS in the recipient. Microbiology. 153(Pt 2):442-51. [PudMed:17259615] |
|
(4) Haft RJ; Palacios G; Nguyen T; Mally M; Gachelet EG; Zechner EL; Traxler B (2006). General mutagenesis of F plasmid TraI reveals its role in conjugative regulation. J Bacteriol. 188(17):6346-53. [PudMed:16923902] |
|
(5) Will WR; Frost LS (2006). Characterization of the opposing roles of H-NS and TraJ in transcriptional regulation of the F-plasmid tra operon. J Bacteriol. 188(2):507-14. [PudMed:16385041] |
|
(6) Will WR; Frost LS (2006). Hfq is a regulator of F-plasmid TraJ and TraM synthesis in Escherichia coli. J Bacteriol. 188(1):124-31. [PudMed:16352828] |
|
(7) Elton TC; Holland SJ; Frost LS; Hazes B (2005). F-like type IV secretion systems encode proteins with thioredoxin folds that are putative DsbC homologues. J Bacteriol. 187(24):8267-77. [PudMed:16321931] |
|
(8) Daehnel K; Harris R; Maddera L; Silverman P (2005). Fluorescence assays for F-pili and their application. Microbiology. 151(Pt 11):3541-8. [PudMed:16272377] |
|
(9) Klimke WA; Rypien CD; Klinger B; Kennedy RA; Rodriguez-Maillard JM; Frost LS (2005). The mating pair stabilization protein, TraN, of the F plasmid is an outer-membrane protein with two regions that are important for its function in conjugation. Microbiology. 151(Pt 11):3527-40. [PudMed:16272376] |
|
(10) Will WR; Lu J; Frost LS (2004). The role of H-NS in silencing F transfer gene expression during entry into stationary phase. Mol Microbiol. 54(3):769-82. [PudMed:15491366] |
|
(11) Harris RL; Silverman PM (2004). Tra proteins characteristic of F-like type IV secretion systems constitute an interaction group by yeast two-hybrid analysis. J Bacteriol. 186(16):5480-5. [PudMed:15292150] |
|
(12) Bidlack JE; Silverman PM (2004). An active type IV secretion system encoded by the F plasmid sensitizes Escherichia coli to bile salts. J Bacteriol. 186(16):5202-9. [PudMed:15292121] |
| (13) Lawley TD; Klimke WA; Gubbins MJ; Frost LS (2003). F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett. 224(1):1-15. [PudMed:12855161] |
|
(14) Schroder G; Krause S; Zechner EL; Traxler B; Yeo HJ; Lurz R; Waksman G; Lanka E (2002). TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates. J Bacteriol. 184(10):2767-79. [PudMed:11976307] |
|
(15) Penfold SS; Simon J; Frost LS (1996). Regulation of the expression of the traM gene of the F sex factor of Escherichia coli. Mol Microbiol. 20(3):549-58. [PudMed:8736534] |
|
(16) Nelson WC; Morton BS; Lahue EE; Matson SW (1993). Characterization of the Escherichia coli F factor traY gene product and its binding sites. J Bacteriol. 175(8):2221-8. [PudMed:8468282] |
|
(17) van Biesen T; Soderbom F; Wagner EG; Frost LS (1993). Structural and functional analyses of the FinP antisense RNA regulatory system of the F conjugative plasmid. Mol Microbiol. 10(1):35-43. [PudMed:7526120] |
|
(18) Fowler T; Taylor L; Thompson R (1983). The control region of the F plasmid transfer operon: DNA sequence of the traJ and traY genes and characterisation of the traY leads to Z promoter. Gene. 26(1):79-89. [PudMed:6368316] |
|
(19) Mullineaux P; Willetts N (1985). Promoters in the transfer region of plasmid F. Basic Life Sci. 30:605-14. [PudMed:3893413] |
|
(20) Gamas P; Caro L; Galas D; Chandler M (1987). Expression of F transfer functions depends on the Escherichia coli integration host factor. Mol Gen Genet. 207(2-3):302-5. [PudMed:3302598] |
|
(21) Silverman PM; Wickersham E; Harris R (1991). Regulation of the F plasmid traY promoter in Escherichia coli by host and plasmid factors. J Mol Biol. 218(1):119-28. [PudMed:2002497] |
|
(22) Silverman PM; Rother S; Gaudin H (1991). Arc and Sfr functions of the Escherichia coli K-12 arcA gene product are genetically and physiologically separable. J Bacteriol. 173(18):5648-52. [PudMed:1885542] |