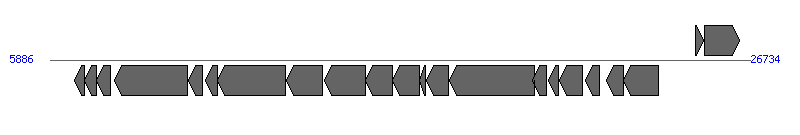The information of T4SS components from NC_011148 |
  |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | SeAg_A0017 | 6629..6922 [-], 294 | 197247331 | hypothetical protein | | |
| 2 | SeAg_A0018 | 6919..7257 [-], 339 | 197247341 | hypothetical protein | | |
| 3 | SeAg_A0019 | 7270..7695 [-], 426 | 197247339 | H-NS histone family | | |
| 4 | SeAg_A0020 | 7793..9973 [-], 2181 | 197247340 | topoisomerase | | |
| 5 | SeAg_A0021 | 10015..10416 [-], 402 | 197247344 | hypothetical protein | | |
| 6 | SeAg_A0022 | 10504..10869 [-], 366 | 197247322 | YggA protein | | |
| 7 | SeAg_A0023 | 10886..12907 [-], 2022 | 197247336 | conjugal transfer protein | VirD4 | |
| 8 | SeAg_A0024 (virB) | 12929..13990 [-], 1062 | 197247321 | P-type DNA transfer ATPase VirB11 | VirB11 | |
| 9 | SeAg_A0025 | 14067..15293 [-], 1227 | 197247337 | conjugal transfer protein | VirB10 | |
| 10 | SeAg_A0026 | 15286..16074 [-], 789 | 197247306 | conjugal transfer outer membrane protein | VirB9 | |
| 11 | SeAg_A0027 | 16096..16887 [-], 792 | 197247349 | conjugal transfer protein | VirB8 | |
| 12 | SeAg_A0028 | 16889..17062 [-], 174 | 197247332 | hypothetical protein | | |
| 13 | SeAg_A0029 | 17112..17765 [-], 654 | 197247313 | putative conjugal transfer protein | VirB5 | |
| 14 | SeAg_A0030 | 17776..20301 [-], 2526 | 197247350 | conjugal transfer protein | VirB4 | |
| 15 | SeAg_A0031 | 20267..20665 [-], 399 | 197247320 | hypothetical protein | | |
| 16 | SeAg_A0032 | 20726..21043 [-], 318 | 197247309 | hypothetical protein | VirB2 | |
| 17 | SeAg_A0033 | 21057..21734 [-], 678 | 197247317 | conjugal transfer protein | VirB1 | |
| 18 | SeAg_A0034 | 21828..22238 [-], 411 | 197247308 | hypothetical protein | | |
| 19 | SeAg_A0035 | 22449..22952 [-], 504 | 197247334 | hypothetical protein | | |
| 20 | SeAg_A0036 | 22953..24017 [-], 1065 | 197247333 | plasmid replication protein | | |
| 21 | SeAg_A0037 | 25110..25358 [+], 249 | 197247315 | hypothetical protein | | |
| 22 | SeAg_A0038 | 25378..26430 [+], 1053 | 197247312 | putative conjugal transfer protein | | |
| |