
| 451 | |
| Escherichia coli plasmid pMAS2027 (NC_013503) | |
| plasmid pMAS2027 [Browse all T4SS(s) in this replicon] | |
| NC_013503 | |
| 23724..35558 | |
| VirB | |
| conjugation | |
| Type IVA; Type P | |
T4SS components |
| Component | VirB1 | VirB2 | VirB3 | VirB4 | VirB5 | VirB6 | VirB7 | VirB8 | VirB9 |
| Number | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| Component | VirB10 | VirB11 | VirD4 | ||||||
| Number | 1 | 1 | 1 |
The information of T4SS components from NC_013503 | ||||||
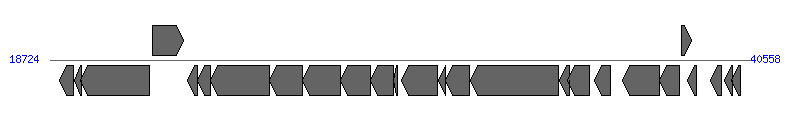 | ||||||
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | pMAS2027_29 | 19008..19469 [-], 462 | 268318489 | DNA binding protein | ||
| 2 | pMAS2027_30 (ydfA) | 19485..19691 [-], 207 | 268318490 | putative hemolysin expression modulating protein | ||
| 3 | pMAS2027_31 (ydgA) | 19688..21841 [-], 2154 | 268318491 | DNA topoisomerase 3 | ||
| 4 | pMAS2027_32 | 21935..22900 [+], 966 | 268318492 | hypothetical protein | ||
| 5 | pMAS2027_33 | 23015..23317 [-], 303 | 268318493 | killer protein | ||
| 6 | pMAS2027_34 | 23314..23727 [-], 414 | 268318494 | putative lipoprotein | ||
| 7 | pMAS2027_35 (taxB) | 23724..25565 [-], 1842 | 268318495 | TaxB | VirD4 | |
| 8 | pMAS2027_36 (pilX11) | 25562..26593 [-], 1032 | 268318496 | PilX11 | VirB11 | |
| 9 | pMAS2027_37 (pilX10) | 26595..27800 [-], 1206 | 268318497 | PilX10 | VirB10 | |
| 10 | pMAS2027_38 (pilX9) | 27797..28726 [-], 930 | 268318498 | PilX9 | VirB9 | |
| 11 | pMAS2027_39 (pilX8) | 28731..29444 [-], 714 | 268318499 | PilX8 | VirB8 | |
| 12 | pMAS2027_40 (pilX7) | 29434..29562 [-], 129 | 268318500 | PilX7 | VirB7 | |
| 13 | pMAS2027_41 (pilX6) | 29689..30822 [-], 1134 | 268318501 | PilX6 | VirB6 | |
| 14 | pMAS2027_42 (eex) | 30832..31059 [-], 228 | 268318502 | EeX | ||
| 15 | pMAS2027_43 (pilX5) | 31060..31818 [-], 759 | 268318503 | PilX5 | VirB5 | |
| 16 | pMAS2027_44 (pilX3-4) | 31829..34582 [-], 2754 | 268318504 | PilX3-4 | VirB4 | |
| 17 | pMAS2027_45 (pilX2) | 34601..34930 [-], 330 | 268318505 | PilX2 | VirB2 | |
| 18 | pMAS2027_46 (pilX1) | 34875..35558 [-], 684 | 268318506 | PilX1 | VirB1 | |
| 19 | pMAS2027_47 (actX) | 35697..36215 [-], 519 | 268318507 | ActX | ||
| 20 | pMAS2027_48 (taxC) | 36568..37740 [-], 1173 | 268318508 | TaxC | ||
| 21 | pMAS2027_49 (ddp1) | 37737..38351 [-], 615 | 268318509 | DNA distortion polypeptide 1 | ||
| 22 | pMAS2027_50 | 38407..38742 [+], 336 | 268318510 | hypothetical protein | ||
| 23 | pMAS2027_51 | 38608..38880 [-], 273 | 268318511 | hypothetical protein | ||
| 24 | pMAS2027_52 (yagA) | 39335..39664 [-], 330 | 268318512 | hypothetical protein | ||
| 25 | pMAS2027_53 (yajA) | 39758..40000 [-], 243 | 268318513 | hypothetical protein | ||
| 26 | pMAS2027_54 | 39990..40271 [-], 282 | 268318514 | hypothetical protein | ||
Download FASTA format files |
| Proteins Genes |
|
(1) Ong CL; Beatson SA; McEwan AG; Schembri MA (2009). Conjugative plasmid transfer and adhesion dynamics in an Escherichia coli biofilm. Appl Environ Microbiol. 75(21):6783-91. [PudMed:19717626] |