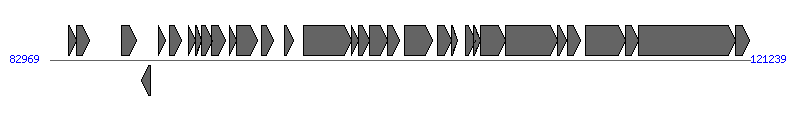The information of T4SS components from NC_009602 |
 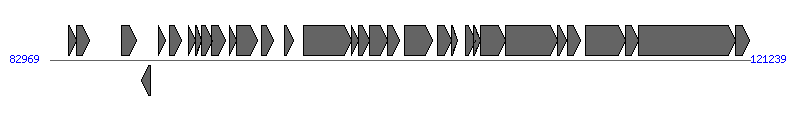 |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | SF0157_p65 (psiB) | 83964..84398 [+], 435 | 149930834 | plasmid SOS inhibition protein B | | |
| 2 | SF0157_p66 | 84395..85114 [+], 720 | 149930816 | plasmid SOS inhibition protein A | | |
| 3 | SF0157_p67 | 86852..87673 [+], 822 | 149930809 | w0067 | | |
| 4 | SF0157_p68 | 87969..88478 [-], 510 | 149930830 | lytic transglycosylase | Orf169_F | |
| 5 | SF0157_p69 | 88910..89293 [+], 384 | 149930839 | conjugal transfer protein TraM | TraM_F |  |
| 6 | SF0157_p70 | 89484..90131 [+], 648 | 149930819 | conjugal transfer transcriptional regulator TraJ | | |
| 7 | SF0157_p71 | 90525..90923 [+], 399 | 149930837 | conjugal transfer pilin subunit TraA | TraA_F | |
| 8 | SF0157_p72 | 90938..91249 [+], 312 | 149930840 | conjugal transfer pilus assembly protein TraL | TraL_F | |
| 9 | SF0157_p73 | 91271..91837 [+], 567 | 149930824 | conjugal transfer pilus assembly protein TraE | TraE_F | |
| 10 | SF0157_p74 | 91824..92552 [+], 729 | 149930814 | conjugal transfer protein TraK | TraK_F | |
| 11 | SF0157_p75 | 92773..93186 [+], 414 | 149930806 | w0075 | | |
| 12 | SF0157_p76 | 93143..94294 [+], 1152 | 149930805 | w0076 | | |
| 13 | SF0157_p77 | 94532..95209 [+], 678 | 149930818 | w0077 | | |
| 14 | SF0157_p78 | 95779..96294 [+], 516 | 149930828 | conjugal transfer protein TraV | TraV_F | |
| 15 | SF0157_p79 | 96810..99437 [+], 2628 | 149930801 | conjugal transfer ATP-binding protein TraC | TraC_F | |
| 16 | SF0157_p80 | 99434..99820 [+], 387 | 149930838 | conjugal transfer protein TrbI | | |
| 17 | SF0157_p81 | 99817..100449 [+], 633 | 149930821 | conjugal transfer pilus assembly protein TraW | TraW_F | |
| 18 | SF0157_p82 | 100446..101438 [+], 993 | 149930802 | conjugal transfer pilus assembly protein TraU | TraU_F | |
| 19 | SF0157_p83 | 101444..102085 [+], 642 | 149930820 | conjugal transfer pilus assembly protein TrbC | TrbC_F | |
| 20 | SF0157_p84 | 102370..103890 [+], 1521 | 149930804 | conjugal transfer mating pair stabilization protein TraN | TraN_F | |
| 21 | SF0157_p85 | 104137..104910 [+], 774 | 149930811 | conjugal pilus assembly protein TraF | TraF_F | |
| 22 | SF0157_p86 | 104926..105273 [+], 348 | 149930842 | conjugal transfer protein TrbA | | |
| 23 | SF0157_p87 | 105663..106208 [+], 546 | 149930827 | conjugal transfer protein TrbB | | |
| 24 | SF0157_p88 | 106138..106500 [+], 363 | 149930843 | conjugal transfer protein TrbJ | | |
| 25 | SF0157_p89 | 106497..107873 [+], 1377 | 149930800 | conjugal transfer pilus assembly protein TraH | TraH_F | |
| 26 | SF0157_p90 | 107870..110692 [+], 2823 | 149930799 | conjugal transfer mating pair stabilization protein TraG | TraG_F | |
| 27 | SF0157_p91 | 110708..111193 [+], 486 | 149930832 | w0091 | | |
| 28 | SF0157_p92 | 111242..111973 [+], 732 | 149930813 | conjugal transfer surface exclusion protein TraT | | |
| 29 | SF0157_p93 | 112226..114433 [+], 2208 | 149930798 | conjugal transfer protein TraD | TraD_F | |
| 30 | SF0157_p94 | 114433..115152 [+], 720 | 149930815 | w0094 | | |
| 31 | SF0157_p95 | 115149..120419 [+], 5271 | 149930845 | conjugal transfer nickase/helicase TraI | TraI_F | |
| 32 | SF0157_p96 | 120439..121185 [+], 747 | 149930812 | conjugal transfer pilus acetylation protein TraX | TraX_F | |
| |