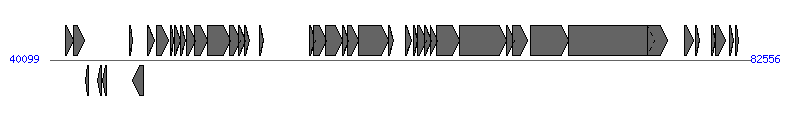The information of T4SS components from NC_014384 |
 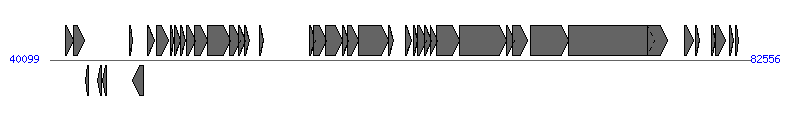 |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | pECL8_p044 (psiB) | 41063..41500 [+], 438 | 302595506 | plasmid SOS inhibition protein B | | |
| 2 | pECL8_p045 (psiA) | 41497..42216 [+], 720 | 302595507 | plasmid SOS inhibition protein A | | |
| 3 | pECL8_p046 (mok) | 42228..42458 [-], 231 | 302595508 | modulator of Hok protein | | |
| 4 | pECL8_p048 | 43008..43232 [-], 225 | 302595509 | hypothetical protein | | |
| 5 | pECL8_p049 | 43276..43509 [-], 234 | 302595510 | hypothetical protein | | |
| 6 | pECL8_p050 (yubP) | 44921..45073 [+], 153 | 302595511 | hypothetical protein | | |
| 7 | pECL8_p051 (geneX) | 45099..45776 [-], 678 | 302595512 | X-polypeptide | Orf169_F | |
| 8 | pECL8_p052 (traM) | 46032..46415 [+], 384 | 302595513 | mating signal | TraM_F |  |
| 9 | pECL8_p053 (traJ) | 46549..47295 [+], 747 | 302595514 | regulation protein | | |
| 10 | pECL8_p054 (traY) | 47389..47616 [+], 228 | 302595515 | OriT nicking | TraY_F | |
| 11 | pECL8_p055 (traA) | 47626..48012 [+], 387 | 302595516 | F pilin subunit | TraA_F | |
| 12 | pECL8_p056 (traL) | 48014..48328 [+], 315 | 302595517 | F pilin assembly | TraL_F | |
| 13 | pECL8_p057 (traE) | 48350..48916 [+], 567 | 302595518 | F pilin assembly | TraE_F | |
| 14 | pECL8_p058 (traK) | 48882..49631 [+], 750 | 302595519 | F pilin assembly | TraK_F | |
| 15 | pECL8_p059 (traB) | 49628..51058 [+], 1431 | 302595520 | F pilin assembly | TraB_F | |
| 16 | pECL8_p060 (traP) | 51006..51632 [+], 627 | 302595521 | conjugal transfer protein | | |
| 17 | pECL8_p061 (trbD) | 51529..51939 [+], 411 | 302595522 | conjugal transfer protein | | |
| 18 | pECL8_p062 (trbG) | 51917..52183 [+], 267 | 302595523 | conjugal transfer protein | | |
| 19 | pECL8_p064 (traR) | 52830..53051 [+], 222 | 302595524 | conjugal transfer protein | | |
| 20 | pECL8_p066 (trbI) | 55820..56221 [+], 402 | 302595525 | conjugal transfer protein | | |
| 21 | pECL8_p067 (traW) | 56104..56850 [+], 747 | 302595526 | F pilus assembly | TraW_F | |
| 22 | pECL8_p068 (traU) | 56820..57839 [+], 1020 | 302595527 | F pilin assembly | TraU_F | |
| 23 | pECL8_p069 (yfdA) | 57863..58174 [+], 312 | 302595528 | hypothetical protein | | |
| 24 | pECL8_p070 (trbC) | 58171..58821 [+], 651 | 302595529 | F pilin assembly | TrbC_F | |
| 25 | pECL8_p071 (traN) | 58818..60626 [+], 1809 | 302595530 | type IV secretion-like conjugative transfer system mating pair stabilization protein | TraN_F | |
| 26 | pECL8_p072 (trbE) | 60650..60910 [+], 261 | 302595531 | conjugal transfer protein | | |
| 27 | pECL8_p074 (trbA) | 61662..62000 [+], 339 | 302595532 | conjugative transfer protein | | |
| 28 | pECL8_p075 (traQ) | 62127..62411 [+], 285 | 302595533 | conjugal transfer protein | TraQ_F | |
| 29 | pECL8_p076 (trbB) | 62398..62943 [+], 546 | 302595534 | F pilin assembly periplasmic protein | | |
| 30 | pECL8_p077 (trbJ) | 62792..63214 [+], 423 | 302595535 | conjugal transfer protein | | |
| 31 | pECL8_p078 (trbF) | 63162..63587 [+], 426 | 302595536 | conjugal transfer protein | | |
| 32 | pECL8_p079 (traH) | 63568..64947 [+], 1380 | 302595537 | F pilin assembly | TraH_F | |
| 33 | pECL8_p080 (traG) | 64944..67763 [+], 2820 | 302595538 | type IV secretion-like conjugative transfer system protein | TraG_F | |
| 34 | pECL8_p081 (traS) | 67782..68279 [+], 498 | 302595539 | conjugal transfer protein | | |
| 35 | pECL8_p082 (traT) | 68161..69042 [+], 882 | 302595540 | conjugal transfer surface exclusion protein | | |
| 36 | pECL8_p083 (traD) | 69268..71520 [+], 2253 | 302595541 | coupling protein | TraD_F | |
| 37 | pECL8_p084 (traI) | 71517..76790 [+], 5274 | 302595542 | nickase/helicase | TraI_F | |
| 38 | pECL8_p085 (traX) | 76339..77556 [+], 1218 | 302595543 | F pilin acetylation protein | TraX_F | |
| 39 | pECL8_p086 (finO) | 78554..79138 [+], 585 | 302595544 | fertility inhibition protein | | |
| 40 | pECL8_p087 (yigA) | 79228..79479 [+], 252 | 302595545 | hypothetical protein | | |
| 41 | pECL8_p089 (hhA) | 80201..80440 [+], 240 | 302595546 | hypothetical protein | | |
| 42 | pECL8_p090 (yihA) | 80478..81068 [+], 591 | 302595547 | hypothetical protein | | |
| 43 | pECL8_p091 (repA2) | 81308..81568 [+], 261 | 302595548 | negative regulator of repA1 expression in FII replicon | | |
| 44 | pECL8_p092 (repA3) | 81653..81847 [+], 195 | 302595549 | regulator of repA1 expression in FII replicon | | |
| |