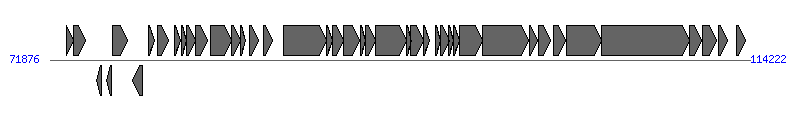The information of T4SS components from NC_013175 |
 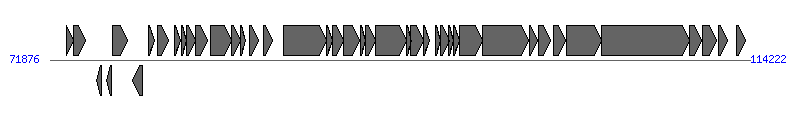 |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | pEC14_76 (psiB) | 72855..73289 [+], 435 | 256855325 | plasmid SOS inhibition protein B | | |
| 2 | pEC14_77 (psiA) | 73286..74005 [+], 720 | 256855326 | plasmid SOS inhibition protein A | | |
| 3 | pEC14_78 | 74676..74999 [-], 324 | 256855327 | hypothetical protein | | |
| 4 | pEC14_79 | 75287..75580 [-], 294 | 256855328 | hypothetical protein | | |
| 5 | pEC14_80 | 75675..76580 [+], 906 | 256855329 | hypothetical protein | | |
| 6 | pEC14_81 | 76876..77466 [-], 591 | 256855330 | lytic transglycosylase | Orf169_F | |
| 7 | pEC14_82 (traM) | 77809..78192 [+], 384 | 256855331 | conjugal transfer protein TraM | TraM_F |  |
| 8 | pEC14_83 (traJ) | 78386..79072 [+], 687 | 256855332 | IncF plasmid conjugative transfer protein TraJ | | |
| 9 | pEC14_84 (traA) | 79427..79786 [+], 360 | 256855333 | conjugal transfer pilin subunit TraA | TraA_F | |
| 10 | pEC14_85 (traL) | 79801..80112 [+], 312 | 256855334 | conjugal transfer pilus assembly protein TraL | TraL_F | |
| 11 | pEC14_86 (traE) | 80134..80700 [+], 567 | 256855335 | conjugal transfer pilus assembly protein TraE | TraE_F | |
| 12 | pEC14_87 (traK) | 80687..81415 [+], 729 | 256855336 | conjugal transfer protein TraK | TraK_F | |
| 13 | pEC14_88 (traB) | 81573..82841 [+], 1269 | 256855337 | IncF plasmid conjugative transfer pilus assembly protein TraB | TraB_F | |
| 14 | pEC14_89 (traP) | 82831..83421 [+], 591 | 256855338 | conjugal transfer protein TraP | | |
| 15 | pEC14_90 (trbD) | 83408..83728 [+], 321 | 256855339 | conjugal transfer protein TrbD | | |
| 16 | pEC14_91 (traV) | 83970..84485 [+], 516 | 256855340 | conjugal transfer protein TraV | TraV_F | |
| 17 | pEC14_92 | 84792..85307 [+], 516 | 256855341 | hypothetical protein | | |
| 18 | pEC14_93 (traC) | 85972..88602 [+], 2631 | 256855342 | conjugal transfer ATP-binding protein TraC | TraC_F | |
| 19 | pEC14_94 (trbI) | 88599..88985 [+], 387 | 256855343 | conjugal transfer protein TrbI | | |
| 20 | pEC14_95 (traW) | 88982..89614 [+], 633 | 256855344 | conjugal transfer pilus assembly protein TraW | TraW_F | |
| 21 | pEC14_96 (traU) | 89611..90603 [+], 993 | 256855345 | conjugal transfer pilus assembly protein TraU | TraU_F | |
| 22 | pEC14_97 | 90633..90938 [+], 306 | 256855346 | hypothetical protein | | |
| 23 | pEC14_98 (trbC) | 90947..91585 [+], 639 | 256855347 | conjugal transfer pilus assembly protein TrbC | TrbC_F | |
| 24 | pEC14_99 (traN) | 91582..93432 [+], 1851 | 256855348 | conjugal transfer mating pair stabilization protein TraN | TraN_F | |
| 25 | pEC14_100 (trbE) | 93459..93716 [+], 258 | 256855349 | conjugal transfer protein TrbE | | |
| 26 | pEC14_101 (traF) | 93709..94452 [+], 744 | 256855350 | conjugal pilus assembly protein TraF | TraF_F | |
| 27 | pEC14_102 (trbA) | 94466..94807 [+], 342 | 256855351 | conjugal transfer protein TrbA | | |
| 28 | pEC14_103 (traQ) | 95204..95488 [+], 285 | 256855352 | conjugal transfer pilin chaperone TraQ | TraQ_F | |
| 29 | pEC14_104 (trbB) | 95475..96020 [+], 546 | 256855353 | conjugal transfer protein TrbB | | |
| 30 | pEC14_105 (trbJ) | 96010..96297 [+], 288 | 256855354 | conjugal transfer protein TrbJ | | |
| 31 | pEC14_106 (trbF) | 96278..96670 [+], 393 | 256855355 | conjugal transfer protein TrbF | | |
| 32 | pEC14_107 (traH) | 96657..98030 [+], 1374 | 256855356 | conjugal transfer pilus assembly protein TraH | TraH_F | |
| 33 | pEC14_108 (traG) | 98027..100849 [+], 2823 | 256855357 | conjugal transfer mating pair stabilization protein TraG | TraG_F | |
| 34 | pEC14_109 (traS) | 100904..101350 [+], 447 | 256855358 | IncF plasmid conjugative transfer surface exclusion protein TraS | | |
| 35 | pEC14_110 (traT) | 101399..102130 [+], 732 | 256855359 | conjugal transfer surface exclusion protein TraT | | |
| 36 | pEC14_111 | 102333..103070 [+], 738 | 256855360 | hypothetical protein | | |
| 37 | pEC14_112 (traD) | 103121..105265 [+], 2145 | 256855361 | conjugal transfer protein TraD | TraD_F | |
| 38 | pEC14_113 (traI) | 105265..110535 [+], 5271 | 256855362 | conjugal transfer nickase/helicase TraI | TraI_F | |
| 39 | pEC14_114 (traX) | 110555..111301 [+], 747 | 256855363 | conjugal transfer pilus acetylation protein TraX | TraX_F | |
| 40 | pEC14_115 | 111360..112220 [+], 861 | 256855364 | hypothetical protein | | |
| 41 | pEC14_116 (finO) | 112323..112883 [+], 561 | 256855365 | conjugal transfer fertility inhibition protein FinO | | |
| 42 | pEC14_117 | 113406..113930 [+], 525 | 256855366 | putative regulatory protein | | |
| |