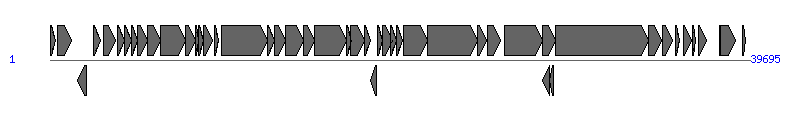The information of T4SS components from NC_004998 |
 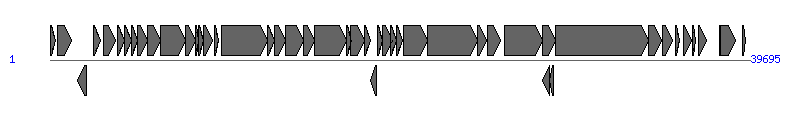 |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | p165897_001 (orf1) | 14..301 [+], 288 | 32469974 | hypothetical protein | | |
| 2 | p165897_002 (orf2) | 423..1244 [+], 822 | 32469975 | hypothetical protein | | |
| 3 | p165897_003 (orf3) | 1541..2050 [-], 510 | 32469976 | lytic transglycosylase | Orf169_F | |
| 4 | p165897_004 (traM) | 2464..2847 [+], 384 | 32469977 | conjugal transfer protein TraM | TraM_F |  |
| 5 | p165897_005 (traJ) | 3034..3723 [+], 690 | 32469978 | conjugal transfer transcriptional regulator TraJ | | |
| 6 | p165897_006 (traY) | 3822..4217 [+], 396 | 32469979 | conjugal transfer protein TraY | TraY_F | |
| 7 | p165897_007 (traA) | 4250..4615 [+], 366 | 32469980 | conjugal transfer pilin subunit TraA | TraA_F | |
| 8 | p165897_008 (traL) | 4630..4941 [+], 312 | 32469981 | conjugal transfer pilus assembly protein TraL | TraL_F | |
| 9 | p165897_009 (traE) | 4963..5529 [+], 567 | 32469982 | conjugal transfer pilus assembly protein TraE | TraE_F | |
| 10 | p165897_010 (traK) | 5516..6244 [+], 729 | 32469983 | conjugal transfer protein TraK | TraK_F | |
| 11 | p165897_011 (traB) | 6244..7671 [+], 1428 | 32469984 | conjugal transfer pilus assembly protein TraB | TraB_F | |
| 12 | p165897_012 (traP) | 7661..8251 [+], 591 | 32469985 | conjugal transfer protein TraP | | |
| 13 | p165897_013 (trbD) | 8238..8435 [+], 198 | 32469986 | conjugal transfer protein TrbD | | |
| 14 | p165897_014 (trbG) | 8447..8698 [+], 252 | 32469987 | conjugal transfer protein TrbG | | |
| 15 | p165897_015 (traV) | 8695..9210 [+], 516 | 32469988 | conjugal transfer protein TraV | TraV_F | |
| 16 | p165897_016 (traR) | 9345..9566 [+], 222 | 32469989 | conjugal transfer protein TraR | | |
| 17 | p165897_017 (traC) | 9726..12353 [+], 2628 | 32469990 | conjugal transfer ATP-binding protein TraC | TraC_F | |
| 18 | p165897_018 (trbI) | 12350..12736 [+], 387 | 32470111 | conjugal transfer protein TrbI | | |
| 19 | p165897_019 (traW) | 12733..13365 [+], 633 | 32469991 | conjugal transfer pilus assembly protein TraW | TraW_F | |
| 20 | p165897_020 (traU) | 13362..14354 [+], 993 | 32469992 | conjugal transfer pilus assembly protein TraU | TraU_F | |
| 21 | p165897_021 (trbC) | 14363..15001 [+], 639 | 32469993 | conjugal transfer pilus assembly protein TrbC | TrbC_F | |
| 22 | p165897_022 (traN) | 14998..16806 [+], 1809 | 32469994 | conjugal transfer mating pair stabilization protein TraN | TraN_F | |
| 23 | p165897_023 (trbE) | 16830..17090 [+], 261 | 32469995 | conjugal transfer protein TrbE | | |
| 24 | p165897_024 (traF) | 17053..17826 [+], 774 | 32469996 | conjugal pilus assembly protein TraF | TraF_F | |
| 25 | p165897_025 (trbA) | 17842..18189 [+], 348 | 32469997 | conjugal transfer protein TrbA | | |
| 26 | p165897_026 (artA) | 18191..18505 [-], 315 | 32469998 | hypothetical protein | | |
| 27 | p165897_027 (traQ) | 18586..18870 [+], 285 | 32469999 | conjugal transfer pilin chaperone TraQ | TraQ_F | |
| 28 | p165897_028 (trbB) | 18857..19402 [+], 546 | 32470000 | conjugal transfer protein TrbB | | |
| 29 | p165897_029 (trbJ) | 19332..19673 [+], 342 | 32470001 | conjugal transfer protein TrbJ | | |
| 30 | p165897_030 (trbF) | 19660..19965 [+], 306 | 32470002 | conjugal transfer protein TrbF | | |
| 31 | p165897_031 (traH) | 20038..21414 [+], 1377 | 32470003 | conjugal transfer pilus assembly protein TraH | TraH_F | |
| 32 | p165897_032 (traG) | 21411..24227 [+], 2817 | 32470004 | conjugal transfer mating pair stabilization protein TraG | TraG_F | |
| 33 | p165897_033 (traS) | 24260..24781 [+], 522 | 32470005 | conjugal transfer entry exclusion protein TraS | | |
| 34 | p165897_034 (traT) | 24803..25537 [+], 735 | 32470006 | conjugal transfer surface exclusion protein TraT | | |
| 35 | p165897_035 (traD) | 25790..27943 [+], 2154 | 32470007 | conjugal transfer protein TraD | TraD_F | |
| 36 | p165897_036 (trbH) | 27943..28662 [+], 720 | 32470008 | TrbH | | |
| 37 | p165897_037 (orf5) | 27952..28350 [-], 399 | 32470009 | plasmid maintenance protein | | |
| 38 | p165897_038 (orf6) | 28350..28577 [-], 228 | 32470010 | hypothetical protein | | |
| 39 | p165897_039 (traI) | 28659..33929 [+], 5271 | 32470011 | conjugal transfer nickase/helicase TraI | TraI_F | |
| 40 | p165897_040 (traX) | 33949..34695 [+], 747 | 32470012 | conjugal transfer pilus acetylation protein TraX | TraX_F | |
| 41 | p165897_041 (finO) | 34750..35310 [+], 561 | 32470013 | conjugal transfer fertility inhibition protein FinO | | |
| 42 | p165897_042 (orf7) | 35452..35706 [+], 255 | 32470014 | hypothetical protein | | |
| 43 | p165897_043 (orfA) | 35903..36364 [+], 462 | 32470015 | hypothetical protein | | |
| 44 | p165897_044 (orf8) | 36410..36619 [+], 210 | 32470016 | putative regulator | | |
| 45 | p165897_045 (orf9) | 36787..37230 [+], 444 | 32470017 | hypothetical protein | | |
| 46 | p165897_046 (repA6) | 37955..38029 [+], 75 | 32470112 | leader peptide RepL | | |
| 47 | p165897_047 (repA1) | 38022..38879 [+], 858 | 32470018 | replication protein | | |
| 48 | p165897_048 (repA4) | 39243..39449 [+], 207 | 32470113 | hypothetical protein | | |
| |