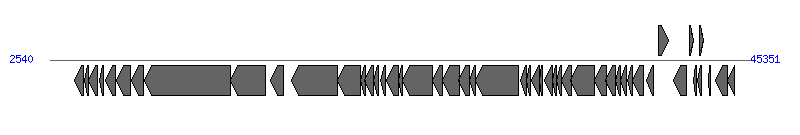The information of T4SS components from NC_006671 |
 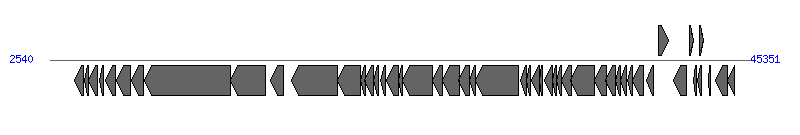 |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | O2R_9 | 4028..4618 [-], 591 | 58000398 | hypothetical protein | | |
| 2 | O2R_10 (hha) | 4656..4865 [-], 210 | 58000397 | Hha protein | | |
| 3 | O2R_11 (yigB) | 4911..5435 [-], 525 | 58000396 | YigB | | |
| 4 | O2R_12 | 5571..5813 [-], 243 | 58000395 | hypothetical protein | | |
| 5 | O2R_13 (finO) | 5958..6518 [-], 561 | 58000394 | conjugal transfer fertility inhibition protein FinO | | |
| 6 | O2R_14 | 6621..7481 [-], 861 | 58000393 | hypothetical protein | | |
| 7 | O2R_15 (traX) | 7540..8286 [-], 747 | 58000392 | conjugal transfer pilus acetylation protein TraX | TraX_F | |
| 8 | O2R_16 (traI) | 8306..13576 [-], 5271 | 58000391 | conjugal transfer nickase/helicase TraI | TraI_F | |
| 9 | O2R_17 (traD) | 13576..15726 [-], 2151 | 58000390 | conjugal transfer protein TraD | TraD_F | |
| 10 | O2R_18 (traT) | 16027..16803 [-], 777 | 58000389 | conjugal transfer surface exclusion protein TraT | | |
| 11 | O2R_20 (traG) | 17308..20130 [-], 2823 | 58000388 | conjugal transfer mating pair stabilization protein TraG | TraG_F | |
| 12 | O2R_21 (traH) | 20127..21503 [-], 1377 | 58000387 | conjugal transfer pilus assembly protein TraH | TraH_F | |
| 13 | O2R_22 (trbJ) | 21500..21862 [-], 363 | 228861735 | conjugal transfer protein TrbJ | | |
| 14 | O2R_23 | 21792..22337 [-], 546 | 58000385 | conjugal transfer protein TrbB | | |
| 15 | O2R_24 (traQ) | 22324..22608 [-], 285 | 58000384 | conjugal transfer pilin chaperone TraQ | TraQ_F | |
| 16 | O2R_25 (trbA) | 22727..23074 [-], 348 | 58000383 | conjugal transfer protein TrbA | | |
| 17 | O2R_26 (traF) | 23090..23833 [-], 744 | 58000382 | conjugal pilus assembly protein TraF | TraF_F | |
| 18 | O2R_27 | 23826..24083 [-], 258 | 58000381 | conjugal transfer protein TrbE | | |
| 19 | O2R_28 (traN) | 24110..25918 [-], 1809 | 58000380 | conjugal transfer mating pair stabilization protein TraN | TraN_F | |
| 20 | O2R_29 (trbC) | 25915..26517 [-], 603 | 58000379 | conjugal transfer pilus assembly protein TrbC | TrbC_F | |
| 21 | O2R_30 (traU) | 26562..27554 [-], 993 | 58000378 | conjugal transfer pilus assembly protein TraU | TraU_F | |
| 22 | O2R_31 (traW) | 27551..28183 [-], 633 | 228861734 | conjugal transfer pilus assembly protein TraW | TraW_F | |
| 23 | O2R_32 (trbI) | 28180..28566 [-], 387 | 58000376 | conjugal transfer protein TrbI | | |
| 24 | O2R_33 (traC) | 28563..31193 [-], 2631 | 58000375 | conjugal transfer ATP-binding protein TraC | TraC_F | |
| 25 | O2R_34 | 31319..31681 [-], 363 | 58000374 | hypothetical protein | | |
| 26 | O2R_35 | 31709..31924 [-], 216 | 58000373 | hypothetical protein | | |
| 27 | O2R_36 | 32004..32477 [-], 474 | 58000372 | hypothetical protein | | |
| 28 | O2R_37 (traR) | 32470..32682 [-], 213 | 58000371 | conjugal transfer protein TraR | | |
| 29 | O2R_38 (traV) | 32761..33276 [-], 516 | 58000370 | conjugal transfer protein TraV | TraV_F | |
| 30 | O2R_39 (trbG) | 33273..33524 [-], 252 | 58000369 | conjugal transfer protein TrbG | | |
| 31 | O2R_40 (trbD) | 33521..33838 [-], 318 | 58000368 | conjugal transfer protein TrbD | | |
| 32 | O2R_41 (traP) | 33835..34407 [-], 573 | 58000367 | conjugal transfer protein TraP | | |
| 33 | O2R_42 (traB) | 34397..35824 [-], 1428 | 58000366 | conjugal transfer pilus assembly protein TraB | TraB_F | |
| 34 | O2R_43 (traK) | 35824..36552 [-], 729 | 58000365 | conjugal transfer protein TraK | TraK_F | |
| 35 | O2R_44 (traE) | 36539..37105 [-], 567 | 58000364 | conjugal transfer pilus assembly protein TraE | TraE_F | |
| 36 | O2R_45 (traL) | 37127..37438 [-], 312 | 58000363 | conjugal transfer pilus assembly protein TraL | TraL_F | |
| 37 | O2R_46 (traA) | 37453..37812 [-], 360 | 58000362 | conjugal transfer pilin subunit TraA | TraA_F | |
| 38 | O2R_47 (traY) | 37846..38166 [-], 321 | 58000361 | conjugal transfer protein TraY | TraY_F | |
| 39 | O2R_48 (traJ) | 38167..38853 [-], 687 | 58000360 | hypothetical protein | | |
| 40 | O2R_49 (traM) | 39044..39427 [-], 384 | 58000359 | conjugal transfer protein TraM | TraM_F |  |
| 41 | O2R_50 | 39758..40351 [+], 594 | 58000358 | lytic transglycosylase | Orf169_F | |
| 42 | O2R_51 | 40648..41469 [-], 822 | 58000357 | hypothetical protein | | |
| 43 | O2R_52 | 41650..41922 [+], 273 | 58000356 | hypothetical protein | | |
| 44 | O2R_53 | 41902..42108 [-], 207 | 58000355 | hypothetical protein | | |
| 45 | O2R_54 | 42144..42356 [-], 213 | 58000354 | hypothetical protein | | |
| 46 | O2R_55 | 42279..42512 [+], 234 | 58000353 | hypothetical protein | | |
| 47 | O2R_56 (hok) | 42798..42956 [-], 159 | 58000352 | small toxic polypeptide | | |
| 48 | O2R_58 (psiA) | 43236..43955 [-], 720 | 58000351 | plasmid SOS inhibition protein A | | |
| 49 | O2R_59 (psiB) | 43952..44386 [-], 435 | 58000350 | plasmid SOS inhibition protein B | | |
| |