|
(1) Guedon G; Libante V; Coluzzi C; Payot S; Leblond-Bourget N (2017). The Obscure World of Integrative and Mobilizable Elements, Highly Widespread Elements that Pirate Bacterial Conjugative Systems. Genes (Basel). 8(11). [PudMed:29165361]
|
|
(2) Bellanger X; Payot S; Leblond-Bourget N; Guedon G (2014). Conjugative and mobilizable genomic islands in bacteria: evolution and diversity. FEMS Microbiol Rev. 38(4):720-60. [PudMed:24372381]
|
|
(3) Ferreira LQ; Avelar KE; Vieira JM; de Paula GR; Colombo AP; Domingues RM; Ferreira MC (2007). Association between the cfxA gene and transposon Tn4555 in Bacteroides distasonis strains and other Bacteroides species. Curr Microbiol. 54(5):348-53. [PudMed:17486409]
|
|
(4) Bass KA; Hecht DW (2002). Isolation and characterization of cLV25, a Bacteroides fragilis chromosomal transfer factor resembling multiple Bacteroides sp. mobilizable transposons. J Bacteriol. 184(7):1895-904. [PudMed:11889096]
|
|
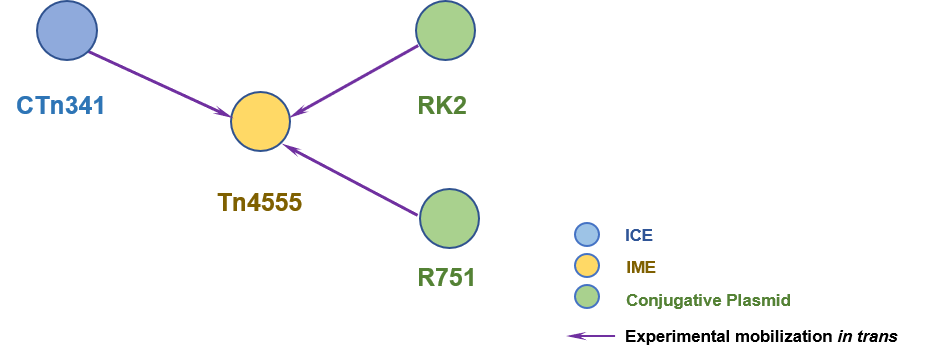
(5) Tribble GD; Parker AC; Smith CJ (1997). The Bacteroides mobilizable transposon Tn4555 integrates by a site-specific recombination mechanism similar to that of the gram-positive bacterial element Tn916. J Bacteriol. 179(8):2731-9. [PudMed:9098073]
|
|
(6) Smith CJ; Parker AC (1996). A gene product related to Tral is required for the mobilization of Bacteroides mobilizable transposons and plasmids. Mol Microbiol. 20(4):741-50. [PudMed:8793871]
|
|
(7) Smith CJ; Parker AC (1993). Identification of a circular intermediate in the transfer and transposition of Tn4555, a mobilizable transposon from Bacteroides spp. J Bacteriol. 175(9):2682-91. [PudMed:8386723]
|