|
(1) Guedon G; Libante V; Coluzzi C; Payot S; Leblond-Bourget N (2017). The Obscure World of Integrative and Mobilizable Elements, Highly Widespread Elements that Pirate Bacterial Conjugative Systems. Genes (Basel). 8(11). [PudMed:29165361]
|
(2) Morici E; Simoni S; Brenciani A; Giovanetti E; Varaldo PE; Mingoia M (2017). A new mosaic integrative and conjugative element from Streptococcus agalactiae carrying resistance genes for chloramphenicol (catQ) and macrolides [mef(I) and erm(TR)]. J Antimicrob Chemother. 72(1):64-67. [PudMed:27621174] 
|
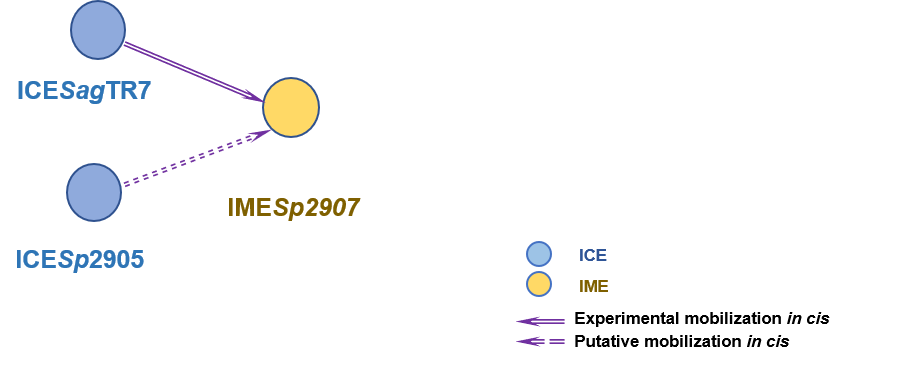
(3) Mingoia M; Morici E; Marini E; Brenciani A; Giovanetti E; Varaldo PE (2016). Macrolide resistance gene erm(TR) and erm(TR)-carrying genetic elements in Streptococcus agalactiae: characterization of ICESagTR7, a new composite element containing IMESp2907. J Antimicrob Chemother. 71(3):593-600. [PudMed:26679245] 
|
|
(4) Bellanger X; Payot S; Leblond-Bourget N; Guedon G (2014). Conjugative and mobilizable genomic islands in bacteria: evolution and diversity. FEMS Microbiol Rev. 38(4):720-60. [PudMed:24372381]
|
(5) Giovanetti E; Brenciani A; Tiberi E; Bacciaglia A; Varaldo PE (2012). ICESp2905, the erm(TR)-tet(O) element of Streptococcus pyogenes, is formed by two independent integrative and conjugative elements. Antimicrob Agents Chemother. 56(1):591-4. [PudMed:21986826] 
|
(6) Brenciani A; Tiberi E; Bacciaglia A; Petrelli D; Varaldo PE; Giovanetti E (2011). Two distinct genetic elements are responsible for erm(TR)-mediated erythromycin resistance in tetracycline-susceptible and tetracycline-resistant strains of Streptococcus pyogenes. Antimicrob Agents Chemother. 55(5):2106-12. [PudMed:21343455] 
|