
| 36 | |
| ICEVflInd1 | |
| ICEO_0000013 | |
| Vibrio fluvialis Ind1 | |
| 114195 | |
| 47.59 | |
| prfC | |
| Antibiotic resistance genes: dfr18, floR, strBA, sul2; Toxin-antitoxin system | |
| - | |
| GQ463144 (complete ICE sequence in this GenBank file) | |
| - | |
| 1..114195 | |
| coordinates: 21141..21247; oriTDB id: 200056 TATCGAGACGCCAAACAGTGATTGTGACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGA TAGTGGTTTTGGCTTTAGGGGTTAATTGGATGGGGAA | |
| - |
The Interaction Network among ICE/IME/CIME | ||||||
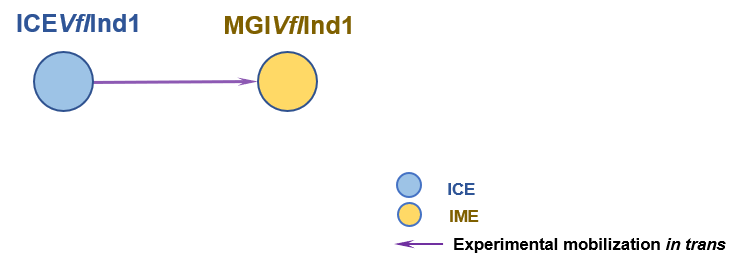 | ||||||
| Detailed Informatioin of the Interaction Network | ||||||
| # | ICE | Inter_Ele | Methods | Donors | Recipients | Exper_Ref |
| 1 | ICEVflInd1 | MGIVflInd1 [IME] | | Vibrio fluvialis; E. coli CAG18439 | Escherichia coli | 20807202; 23204461 |
The graph information of ICEVflInd1 components from GQ463144 | |||||
 | |||||
| Complete gene list of ICEVflInd1 from GQ463144 | |||||
| # | Gene | Coordinates [+/-], size (bp) | Product (GenBank annotation) | *Reannotation | |
| 1 | ICEVFLIND1_0001 | 20..154 [+], 135 | ribosome-binding factor A | ||
| 2 | ICEVFLIND1_0002 | 154..1092 [+], 939 | tRNA pseudouridine synthase B | ||
| 3 | rpsO | 1234..1503 [+], 270 | ribosomal protein S15 | ||
| 4 | pnp | 1866..3992 [+], 2127 | polyribonucleotide nucleotidyltransferase | ||
| 5 | ICEVFLIND1_0005 | 4026..4169 [-], 144 | hypothetical protein | ||
| 6 | ICEVFLIND1_0008 | 5086..5526 [+], 441 | transcriptional regulator, MarR family | ||
| 7 | ICEVFLIND1_0009 | 5569..6906 [+], 1338 | multidrug resistance pump | ||
| 8 | yhbV | 6965..7843 [-], 879 | peptidase, U32 family | ||
| 9 | ICEVFLIND1_0011 | 7855..8868 [-], 1014 | STM-proteaseA | ||
| 10 | ICEVFLIND1_0014 | 11036..11563 [+], 528 | sterol binding protein | ||
| 11 | ICEVFLIND1_0015 | 11547..12050 [+], 504 | acetyltransferase, gnat family | ||
| 12 | ICEVFLIND1_0016 | 12191..12568 [+], 378 | DNA polymerase III, psi subunit | ||
| 13 | rimI | 12565..13014 [+], 450 | ribosomal-protein-alanine acetyltransferase | ||
| 14 | ICEVFLIND1_0017 | 13003..15060 [-], 2058 | ggdef domain protein | ||
| 15 | ICEVFLIND1_0019 | 15852..15989 [+], 138 | conserved domain protein | ||
| 16 | ICEVFLIND1_0022 | 18282..19523 [-], 1242 | integrase | Integrase | |
| 17 | ICEVFLIND1_0023 | 19525..19794 [-], 270 | conserved hypothetical protein | ||
| 18 | ICEVFLIND1_0024 | 19797..20771 [-], 975 | conserved hypothetical protein | ||
| 19 | ICEVFLIND1_0025 | 20867..21034 [+], 168 | hypothetical protein | ||
| 20 | ICEVFLIND1_0026 | 21079..21213 [+], 135 | hypothetical protein | ||
| 21 | ICEVFLIND1_0027 | 21236..21679 [+], 444 | conserved hypothetical protein | ||
| 22 | ICEVFLIND1_0028 | 21690..22862 [-], 1173 | protein UmuC | ||
| 23 | ICEVFLIND1_0029 | 23146..23739 [+], 594 | transposase, IS3 family, interruption | ||
| 24 | ICEVFLIND1_0030 | 23853..26831 [+], 2979 | transposase | ||
| 25 | ICEVFLIND1_0031 | 27249..28742 [+], 1494 | iscr2 transposase | ||
| 26 | ICEVFLIND1_0032 | 28970..29113 [-], 144 | hypothetical protein | ||
| 27 | folA | 29103..29657 [-], 555 | dihydrofolate reductase | ||
| 28 | ICEVFLIND1_0034 | 29671..30156 [-], 486 | conserved hypothetical protein | ||
| 29 | ICEVFLIND1_0035 | 30216..31382 [-], 1167 | conserved hypothetical protein | ||
| 30 | dcd | 31397..32023 [-], 627 | deoxycytidine triphosphate deaminase | ||
| 31 | ICEVFLIND1_0037 | 32316..32717 [+], 402 | transposase | ||
| 32 | ICEVFLIND1_0039 | 33826..35040 [+], 1215 | florfenicol/chloramphenicol resistance protein | AR | |
| 33 | ICEVFLIND1_0040 | 35068..35373 [+], 306 | truncated helix-turn-helix multiple antibiotic resistance protein | ||
| 34 | ICEVFLIND1_0042 | 35485..36024 [+], 540 | truncated transposase | ||
| 35 | ICEVFLIND1_0041 | 35996..36826 [-], 831 | streptomycin phosphotransferase B | AR | |
| 36 | ICEVFLIND1_0043 | 36832..37635 [-], 804 | streptomycin 3''-kinase | AR | |
| 37 | folP | 37696..38511 [-], 816 | dihydropteroate synthase | AR | |
| 38 | ICEVFLIND1_0045 | 38982..40355 [+], 1374 | transposase Tn3 family protein | ||
| 39 | ICEVFLIND1_0046 | 40940..41209 [-], 270 | protein UmuC | ||
| 40 | ICEVFLIND1_0047 | 41217..41666 [-], 450 | protein UmuD | ||
| 41 | ICEVFLIND1_0048 | 41780..41923 [+], 144 | hypothetical protein | ||
| 42 | ICEVFLIND1_0049 | 42170..42328 [-], 159 | hypothetical protein | ||
| 43 | ICEVFLIND1_0050 | 42307..43212 [+], 906 | exonuclease, RNase T and DNA polymerase III | ||
| 44 | ICEVFLIND1_0051 | 43300..43599 [+], 300 | conserved hypothetical protein | ||
| 45 | ICEVFLIND1_0052 | 43917..44840 [+], 924 | conserved hypothetical protein | ||
| 46 | ICEVFLIND1_0053 | 44954..47383 [+], 2430 | N-6 DNA methylase | ||
| 47 | ICEVFLIND1_0054 | 47383..48552 [+], 1170 | restriction modification system DNA specificity domain | ||
| 48 | ICEVFLIND1_0055 | 48563..49720 [+], 1158 | conserved hypothetical protein | ||
| 49 | ICEVFLIND1_0058 | 51591..51719 [+], 129 | conjugative relaxase | ||
| 50 | ICEVFLIND1_0059 | 51768..53153 [+], 1386 | conjugative coupling factor | TraD_F, T4SS component | |
| 51 | ICEVFLIND1_0060 | 53304..53420 [+], 117 | hypothetical protein | ||
| 52 | ICEVFLIND1_0061 | 53436..54953 [+], 1518 | putative transposase for insertion sequence | ||
| 53 | ICEVFLIND1_0062 | 54977..55672 [+], 696 | conserved hypothetical protein | ||
| 54 | ICEVFLIND1_0063 | 55733..56443 [+], 711 | transposition helper protein | ||
| 55 | ICEVFLIND1_0064 | 56480..57043 [-], 564 | conserved hypothetical protein | ||
| 56 | traL | 57332..57613 [+], 282 | type IV conjugative transfer system protein TraL | TraL_F, T4SS component | |
| 57 | ICEVFLIND1_0066 | 57610..58236 [+], 627 | sex pilus assembly | TraE_F, T4SS component | |
| 58 | ICEVFLIND1_0067 | 58220..59116 [+], 897 | TraK | TraK_F, T4SS component | |
| 59 | ICEVFLIND1_0070 | 60491..61051 [+], 561 | sex pilus assembly | TraV_F, T4SS component | |
| 60 | ICEVFLIND1_0071 | 61048..61434 [+], 387 | putative pilin subunit | TraA_F, T4SS component | |
| 61 | ICEVFLIND1_0072 | 61612..62445 [+], 834 | conserved hypothetical protein | ||
| 62 | ICEVFLIND1_0075 | 63506..64198 [+], 693 | DsbC | TrbB_I, T4SS component | |
| 63 | traC | 64198..66597 [+], 2400 | type-IV secretion system protein TraC | TraC_F, T4SS component | |
| 64 | ICEVFLIND1_0077 | 66656..66937 [+], 282 | conserved hypothetical protein | ||
| 65 | ICEVFLIND1_0078 | 66921..67433 [+], 513 | conjugation signal peptidase | TraF, T4SS component | |
| 66 | ICEVFLIND1_0079 | 67444..68568 [+], 1125 | sex pilus assembly | TraW_F, T4SS component | |
| 67 | ICEVFLIND1_0080 | 68546..69580 [+], 1035 | conjugal transfer protein TraU, putative | TraU_F, T4SS component | |
| 68 | ICEVFLIND1_0081 | 69583..73275 [+], 3693 | mating pair stabilization | TraN_F, T4SS component | |
| 69 | ICEVFLIND1_0082 | 73506..73838 [+], 333 | hypothetical protein | ||
| 70 | ICEVFLIND1_0083 | 73869..74531 [+], 663 | conserved hypothetical protein | ||
| 71 | ICEVFLIND1_0084 | 74622..75224 [-], 603 | conserved hypothetical protein | ||
| 72 | ICEVFLIND1_0085 | 75634..75918 [+], 285 | conserved hypothetical protein | ||
| 73 | ICEVFLIND1_0086 | 75934..76353 [+], 420 | single-strand binding protein family | ||
| 74 | ICEVFLIND1_0087 | 76433..77251 [+], 819 | phage recombination protein Bet | ||
| 75 | ICEVFLIND1_0088 | 77333..77476 [+], 144 | hypothetical protein | ||
| 76 | ICEVFLIND1_0089 | 77537..78553 [+], 1017 | YqaJ viral recombinase family | ||
| 77 | ICEVFLIND1_0090 | 78763..79722 [+], 960 | ATPase associated with various cellular activities, AAA_5 | ||
| 78 | ICEVFLIND1_0091 | 79722..80489 [+], 768 | conserved hypothetical protein | ||
| 79 | ICEVFLIND1_0092 | 80588..81541 [+], 954 | conserved hypothetical protein | ||
| 80 | ICEVFLIND1_0093 | 81603..82043 [+], 441 | conserved hypothetical protein | ||
| 81 | ICEVFLIND1_0094 | 82113..83768 [+], 1656 | von Willebrand factor, type A | ||
| 82 | ICEVFLIND1_0095 | 83851..84348 [+], 498 | DNA repair protein RadC | ||
| 83 | ICEVFLIND1_0096 | 84348..84689 [+], 342 | conserved hypothetical protein | ||
| 84 | ICEVFLIND1_0097 | 84781..85854 [+], 1074 | phage P4 alpha, zinc-binding domain protein | ||
| 85 | ICEVFLIND1_0098 | 85947..86651 [+], 705 | conserved hypothetical protein | ||
| 86 | ICEVFLIND1_0099 | 86674..86799 [+], 126 | hypothetical protein | ||
| 87 | ICEVFLIND1_0100 | 86768..87520 [+], 753 | conserved hypothetical protein | ||
| 88 | ICEVFLIND1_0101 | 87575..88006 [-], 432 | conserved hypothetical protein | ||
| 89 | ICEVFLIND1_0102 | 88044..88163 [+], 120 | hypothetical protein | ||
| 90 | ICEVFLIND1_0103 | 88472..89743 [+], 1272 | pas/pac/ggdef-domain containing protein | ||
| 91 | ICEVFLIND1_0104 | 89917..90342 [+], 426 | integrase, catalytic region | ||
| 92 | ICEVFLIND1_0105 | 90503..91432 [-], 930 | response regulator receiver protein | ||
| 93 | ICEVFLIND1_0106 | 91438..95196 [-], 3759 | multi-sensor hybrid histidine kinase | ||
| 94 | ICEVFLIND1_0107 | 95245..95361 [-], 117 | hypothetical protein | ||
| 95 | ICEVFLIND1_0108 | 95680..95838 [+], 159 | hypothetical protein | ||
| 96 | ICEVFLIND1_0109 | 95984..96928 [+], 945 | conserved hypothetical protein | TraF_F, T4SS component | |
| 97 | ICEVFLIND1_0110 | 96931..98319 [+], 1389 | TraH family protein | TraH_F, T4SS component | |
| 98 | ICEVFLIND1_0111 | 98323..101892 [+], 3570 | sex pilus assembly | TraG_F, T4SS component | |
| 99 | ICEVFLIND1_0112 | 101925..102380 [-], 456 | EexS1 | ||
| 100 | ICEVFLIND1_0113 | 102414..102947 [-], 534 | transcriptional activator | ||
| 101 | ICEVFLIND1_0114 | 102944..103243 [-], 300 | transcriptional activator | ||
| 102 | ICEVFLIND1_0115 | 103240..103788 [-], 549 | lytic transglycosylase, catalytic | Orf169_F, T4SS component | |
| 103 | ICEVFLIND1_0116 | 103775..104437 [-], 663 | conserved hypothetical protein | ||
| 104 | ICEVFLIND1_0117 | 104424..105293 [-], 870 | conserved hypothetical protein | ||
| 105 | ICEVFLIND1_0118 | 105349..105600 [-], 252 | conserved hypothetical protein | ||
| 106 | ICEVFLIND1_0119 | 105718..106365 [+], 648 | HTH-type transcriptional regulator for conjugative element R391 | ||
| 107 | prfC | 106622..108211 [+], 1590 | peptide chain release factor 3 | ||
| 108 | ICEVFLIND1_0121 | 108333..109604 [-], 1272 | ATP-dependent RNA helicase SrmB | ||
| 109 | ICEVFLIND1_0122 | 109692..110414 [-], 723 | methyltransferase | ||
| 110 | brnQ | 110723..112036 [+], 1314 | branched-chain amino acid transport system II carrier protein | ||
| 111 | ICEVFLIND1_0125 | 113335..114195 [+], 861 | lysyl-tRNA synthetase (Lysine--tRNA ligase) (LysRS) | ||
|
(1) Daccord A; Mursell M; Poulin-Laprade D; Burrus V (2012). Dynamics of the SetCD-Regulated Integration and Excision of Genomic Islands Mobilized by Integrating Conjugative Elements of the SXT/R391 Family. J Bacteriol. 194(21):5794-802. [PubMed:22923590] |
|
(2) Daccord A; Ceccarelli D; Burrus V (2010). Integrating conjugative elements of the SXT/R391 family trigger the excision and drive the mobilization of a new class of Vibrio genomic islands. Mol Microbiol. 78(3):576-88. [PubMed:20807202] |
|
(3) Wozniak RA; Fouts DE; Spagnoletti M; Colombo MM; Ceccarelli D; Garriss G; Dery C; Burrus V; Waldor MK (2009). Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet. 5(12):e1000786. [PubMed:20041216] |
|
(4) Bordeleau E; Brouillette E; Robichaud N; Burrus V (2010). Beyond antibiotic resistance: integrating conjugative elements of the SXT/R391 family that encode novel diguanylate cyclases participate to c-di-GMP signalling in Vibrio cholerae. Environ Microbiol. 12(2):510-23. [PubMed:19888998] |
|
(5) Marrero J; Waldor MK (2007). The SXT/R391 family of integrative conjugative elements is composed of two exclusion groups. J Bacteriol. 189(8):3302-5. [PubMed:17307849] |