
| 22 | |
| ICEVchInd5 [ICEVchHai1] | |
| ICEO_0000010 | |
| Vibrio cholerae O1 Ind5 | |
| 97952 | |
| 46.73 | |
| prfC | |
| Antibiotic resistance genes: floR, strBA, sul2, dfrA1; glyoxoylase abx resistance | |
| - | |
| GQ463142 (complete ICE sequence in this GenBank file) | |
| - | |
| 1..97952 | |
| coordinates: 5906..6012; oriTDB id: 200056 TATCGAGACGCCAAACAGTGATTGTGACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGA TAGTGGTTTTGGCTTTAGGGGTTAATTGGATGGGGAA | |
| coordinates: 41932..44082; Locus tag: ICEVCHIND5_0038; Family: MOBH |
The graph information of ICEVchInd5 [ICEVchHai1] components from GQ463142 | |||||
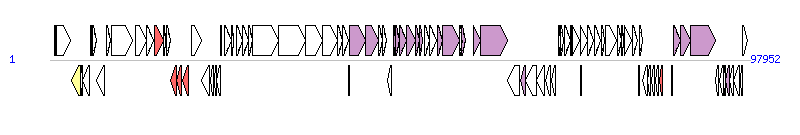 | |||||
| Complete gene list of ICEVchInd5 [ICEVchHai1] from GQ463142 | |||||
| # | Gene | Coordinates [+/-], size (bp) | Product (GenBank annotation) | *Reannotation | |
| 1 | ICEVCHIND5_0001 | 565..753 [+], 189 | conserved domain protein | ||
| 2 | ICEVCHIND5_0002 | 899..2917 [+], 2019 | putative DNA mismatch repair protein | ||
| 3 | ICEVCHIND5_0003 | 3047..4288 [-], 1242 | integrase | Integrase | |
| 4 | ICEVCHIND5_0004 | 4290..4559 [-], 270 | conserved hypothetical protein | ||
| 5 | ICEVCHIND5_0005 | 4562..5536 [-], 975 | conserved hypothetical protein | ||
| 6 | ICEVCHIND5_0006 | 5632..5799 [+], 168 | hypothetical protein | ||
| 7 | ICEVCHIND5_0007 | 5844..5978 [+], 135 | hypothetical protein | ||
| 8 | ICEVCHIND5_0008 | 6001..6444 [+], 444 | conserved hypothetical protein | ||
| 9 | ICEVCHIND5_0009 | 6455..7627 [-], 1173 | protein UmuC | ||
| 10 | ICEVCHIND5_0010 | 7911..8504 [+], 594 | transposase, IS3 family, interruption | ||
| 11 | ICEVCHIND5_0011 | 8618..11596 [+], 2979 | transposase | ||
| 12 | ICEVCHIND5_0012 | 12014..13507 [+], 1494 | iscr2 transposase | ||
| 13 | ICEVCHIND5_0013 | 13538..14422 [+], 885 | conserved hypothetical protein | ||
| 14 | ICEVCHIND5_0014 | 14639..15853 [+], 1215 | florfenicol/chloramphenicol resistance protein | AR | |
| 15 | ICEVCHIND5_0015 | 15908..16186 [+], 279 | truncated helix-turn-helix multiple antibiotic resistance protein | ||
| 16 | ICEVCHIND5_0017 | 16298..16837 [+], 540 | truncated transposase | ||
| 17 | ICEVCHIND5_0016 | 16809..17639 [-], 831 | streptomycin resistance protein B | AR | |
| 18 | ICEVCHIND5_0018 | 17645..18448 [-], 804 | streptomycin 3''-kinase | AR | |
| 19 | folP | 18509..19324 [-], 816 | dihydropteroate synthase | AR | |
| 20 | ICEVCHIND5_0021 | 19795..21147 [+], 1353 | TnpA | ||
| 21 | ICEVCHIND5_0020 | 21139..22350 [-], 1212 | transposase, Mutator family | ||
| 22 | ICEVCHIND5_0022 | 22408..22911 [-], 504 | MutL protein | ||
| 23 | ICEVCHIND5_0023 | 23064..23333 [-], 270 | ultraviolet light resistance protein B | ||
| 24 | ICEVCHIND5_0024 | 23341..23790 [-], 450 | protein UmuD | ||
| 25 | ICEVCHIND5_0025 | 23909..24046 [+], 138 | hypothetical protein | ||
| 26 | ICEVCHIND5_0026 | 24430..25335 [+], 906 | exonuclease, RNase T and DNA polymerase III | ||
| 27 | ICEVCHIND5_0027 | 25423..25722 [+], 300 | conserved hypothetical protein | ||
| 28 | ICEVCHIND5_0028 | 26040..26960 [+], 921 | conserved hypothetical protein | ||
| 29 | ICEVCHIND5_0029 | 26978..27778 [+], 801 | hypothetical protein | ||
| 30 | ICEVCHIND5_0030 | 27762..28310 [+], 549 | conserved hypothetical protein | ||
| 31 | ICEVCHIND5_0031 | 28323..32012 [+], 3690 | conserved hypothetical protein | ||
| 32 | ICEVCHIND5_0032 | 32017..35757 [+], 3741 | conserved hypothetical protein | ||
| 33 | ICEVCHIND5_0033 | 35770..38073 [+], 2304 | PglZ domain family | ||
| 34 | ICEVCHIND5_0034 | 38112..40163 [+], 2052 | conserved hypothetical protein | ||
| 35 | ICEVCHIND5_0035 | 40190..40960 [+], 771 | conserved hypothetical protein | ||
| 36 | ICEVCHIND5_0037 | 40995..41810 [+], 816 | conserved hypothetical protein | ||
| 37 | ICEVCHIND5_0036 | 41787..41909 [-], 123 | hypothetical protein | ||
| 38 | ICEVCHIND5_0038 | 41932..44082 [+], 2151 | conjugative relaxase | Relaxase, MOBH Family | |
| 39 | ICEVCHIND5_0039 | 44131..45951 [+], 1821 | conjugative coupling factor | TraD_F, T4SS component | |
| 40 | ICEVCHIND5_0040 | 45961..46521 [+], 561 | conserved hypothetical protein | ||
| 41 | ICEVCHIND5_0041 | 46508..47143 [+], 636 | putative conjugation coupling factor | ||
| 42 | ICEVCHIND5_0042 | 47170..47757 [-], 588 | conserved hypothetical protein | ||
| 43 | traL | 48046..48327 [+], 282 | type IV conjugative transfer system protein TraL | TraL_F, T4SS component | |
| 44 | ICEVCHIND5_0044 | 48324..48950 [+], 627 | sex pilus assembly | TraE_F, T4SS component | |
| 45 | ICEVCHIND5_0045 | 48934..49830 [+], 897 | TraK | TraK_F, T4SS component | |
| 46 | ICEVCHIND5_0046 | 49833..51122 [+], 1290 | sex pilus assembly | TraB_F, T4SS component | |
| 47 | ICEVCHIND5_0047 | 51119..51769 [+], 651 | sex pilus assembly | TraV_F, T4SS component | |
| 48 | ICEVCHIND5_0048 | 51766..52152 [+], 387 | TraA | TraA_F, T4SS component | |
| 49 | ICEVCHIND5_0049 | 52373..53164 [+], 792 | conserved hypothetical protein | ||
| 50 | ICEVCHIND5_0050 | 53157..54095 [+], 939 | ync | ||
| 51 | ICEVCHIND5_0051 | 54227..54919 [+], 693 | DsbC | TrbB_I, T4SS component | |
| 52 | traC | 54919..57318 [+], 2400 | type-IV secretion system protein TraC | TraC_F, T4SS component | |
| 53 | ICEVCHIND5_0053 | 57311..57658 [+], 348 | conserved hypothetical protein | ||
| 54 | ICEVCHIND5_0054 | 57642..58154 [+], 513 | conjugation signal peptidase | TraF, T4SS component | |
| 55 | ICEVCHIND5_0056 | 59273..60301 [+], 1029 | sex pilus assembly | TraU_F, T4SS component | |
| 56 | ICEVCHIND5_0057 | 60304..63996 [+], 3693 | mating pair stabilization | TraN_F, T4SS component | |
| 57 | ICEVCHIND5_0058 | 64087..65703 [-], 1617 | conserved hypothetical protein | ||
| 58 | ICEVCHIND5_0059 | 65783..66538 [-], 756 | IstB domain protein ATP-binding protein | TrbB, T4SS component | |
| 59 | ICEVCHIND5_0060 | 66528..68036 [-], 1509 | integrase, catalytic region | ||
| 60 | ICEVCHIND5_0061 | 68135..69178 [-], 1044 | SMC domain protein | ||
| 61 | ICEVCHIND5_0062 | 69382..70065 [-], 684 | endonuclease-1 (Endonuclease I) (Endo I) | ||
| 62 | ICEVCHIND5_0063 | 70174..70776 [-], 603 | conserved hypothetical protein | ||
| 63 | ICEVCHIND5_0064 | 71144..71470 [+], 327 | conserved hypothetical protein | ||
| 64 | ICEVCHIND5_0065 | 71486..71905 [+], 420 | single-strand binding protein family | ||
| 65 | ICEVCHIND5_0066 | 71985..72803 [+], 819 | phage recombination protein Bet | ||
| 66 | ICEVCHIND5_0067 | 72885..73028 [+], 144 | hypothetical protein | ||
| 67 | ICEVCHIND5_0068 | 73089..74105 [+], 1017 | YqaJ viral recombinase family | ||
| 68 | ICEVCHIND5_0070 | 74198..75274 [+], 1077 | ATPase associated with various cellular activities, AAA_5 | ||
| 69 | ICEVCHIND5_0069 | 74294..74416 [-], 123 | hypothetical protein | ||
| 70 | ICEVCHIND5_0071 | 75274..76041 [+], 768 | conserved hypothetical protein | ||
| 71 | ICEVCHIND5_0072 | 76140..77093 [+], 954 | conserved hypothetical protein | ||
| 72 | ICEVCHIND5_0073 | 77155..77595 [+], 441 | conserved hypothetical protein | ||
| 73 | ICEVCHIND5_0074 | 77665..79320 [+], 1656 | von Willebrand factor, type A | ||
| 74 | radC | 79405..79902 [+], 498 | DNA repair protein RadC | ||
| 75 | ICEVCHIND5_0076 | 79902..80243 [+], 342 | conserved hypothetical protein | ||
| 76 | ICEVCHIND5_0077 | 80334..81407 [+], 1074 | phage P4 alpha, zinc-binding domain protein | ||
| 77 | ICEVCHIND5_0078 | 81497..82204 [+], 708 | conserved hypothetical protein | ||
| 78 | ICEVCHIND5_0079 | 82397..82510 [-], 114 | hypothetical protein | ||
| 79 | ICEVCHIND5_0081 | 82476..82880 [+], 405 | glyoxalase/bleomycin resistance protein/dioxygenase | ||
| 80 | ICEVCHIND5_0080 | 82877..83578 [-], 702 | conserved hypothetical protein | ||
| 81 | ICEVCHIND5_0082 | 83678..84040 [-], 363 | conserved hypothetical protein | ||
| 82 | ICEVCHIND5_0083 | 84111..84521 [-], 411 | membrane associated protein containing xre-family DNA-binding HTH domain | ||
| 83 | ICEVCHIND5_0084 | 84719..85144 [-], 426 | conserved hypothetical protein | ||
| 84 | folA | 85249..85722 [-], 474 | dihydrofolate reductase | AR | |
| 85 | ICEVCHIND5_0086 | 86940..87080 [-], 141 | hypothetical protein | ||
| 86 | ICEVCHIND5_0088 | 87226..88170 [+], 945 | conserved hypothetical protein | TraF_F, T4SS component | |
| 87 | ICEVCHIND5_0089 | 88173..89561 [+], 1389 | sex pilus assembly | TraH_F, T4SS component | |
| 88 | ICEVCHIND5_0090 | 89565..93134 [+], 3570 | sex pilus assembly | TraG_F, T4SS component | |
| 89 | ICEVCHIND5_0091 | 93167..93598 [-], 432 | EexR1 | ||
| 90 | ICEVCHIND5_0092 | 93656..94189 [-], 534 | transcriptional activator | ||
| 91 | ICEVCHIND5_0093 | 94186..94485 [-], 300 | SetD | ||
| 92 | ICEVCHIND5_0094 | 94482..95030 [-], 549 | lytic transglycosylase, catalytic | Orf169_F, T4SS component | |
| 93 | ICEVCHIND5_0095 | 95017..95679 [-], 663 | conserved hypothetical protein | ||
| 94 | ICEVCHIND5_0096 | 95666..96535 [-], 870 | conserved hypothetical protein | ||
| 95 | ICEVCHIND5_0097 | 96591..96842 [-], 252 | SetQ | ||
| 96 | ICEVCHIND5_0098 | 96960..97607 [+], 648 | HTH-type transcriptional regulator for conjugative element R391 | ||
| (1) Carraro N; Rivard N; Ceccarelli D; Colwell RR; Burrus V (2016). IncA/C Conjugative Plasmids Mobilize a New Family of Multidrug Resistance Islands in Clinical Vibrio cholerae Non-O1/Non-O139 Isolates from Haiti. MBio. 7(4). [PubMed:27435459] |
| (2) Sjolund-Karlsson M; Reimer A; Folster JP; Walker M; Dahourou GA; Batra DG; Martin I; Joyce K; Parsons MB; Boncy J; Whichard JM; Gilmour MW (2011). Drug-resistance mechanisms in Vibrio cholerae O1 outbreak strain, Haiti, 2010. Emerg Infect Dis. 17(11):2151-4. [PubMed:22099122] |
|
(3) Spagnoletti M; Ceccarelli D; Colombo MM (2012). Rapid detection by multiplex PCR of Genomic Islands, prophages and Integrative Conjugative Elements in V. cholerae 7th pandemic variants. J Microbiol Methods. 88(1):98-102. [PubMed:22062086] |
|
(4) Ceccarelli D; Spagnoletti M; Bacciu D; Danin-Poleg Y; Mendiratta DK; Kashi Y; Cappuccinelli P; Burrus V; Colombo MM (2011). ICEVchInd5 is prevalent in epidemic Vibrio cholerae O1 El Tor strains isolated in India. Int J Med Microbiol. 301(4):318-24. [PubMed:21276749] |
|
(5) Wozniak RA; Fouts DE; Spagnoletti M; Colombo MM; Ceccarelli D; Garriss G; Dery C; Burrus V; Waldor MK (2009). Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet. 5(12):e1000786. [PubMed:20041216] |
|
(6) Bordeleau E; Brouillette E; Robichaud N; Burrus V (2010). Beyond antibiotic resistance: integrating conjugative elements of the SXT/R391 family that encode novel diguanylate cyclases participate to c-di-GMP signalling in Vibrio cholerae. Environ Microbiol. 12(2):510-23. [PubMed:19888998] |