The graph information of ICEKp1 components from AP006725 |
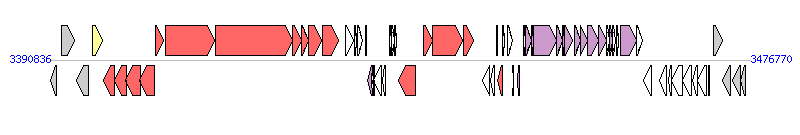 |
| Complete gene list of ICEKp1 from AP006725 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | KP1_3577 | 3390903..3391607 [-], 705 | putative TetR-family trancriptional regulatory protein | |
| 2 | KP1_3578 | 3392198..3393832 [+], 1635 | putative sodium:hydrogen antiporter | |
| 3 | yeeO | 3394131..3395600 [-], 1470 | putative MATE family transport protein | |
| 4 | int | 3395997..3397259 [+], 1263 | integrase | Integrase |
| 5 | ybtS | 3397453..3398757 [-], 1305 | putative salicylate synthetase | VF |
| 6 | ybtX | 3398785..3400188 [-], 1404 | putative cytoplasmic transmembrane protein | VF |
| 7 | ybtQ | 3400058..3401887 [-], 1830 | putative ABC transporter protein | VF |
| 8 | ybtP | 3401847..3403649 [-], 1803 | putative inner membrane ABC-transporter | VF |
| 9 | ybtA | 3403816..3404775 [+], 960 | transcriptional regulator | VF |
| 10 | irp2 | 3404966..3411073 [+], 6108 | yersiniabactin biosynthetic protein | VF |
| 11 | irp1 | 3411161..3420652 [+], 9492 | yersiniabactin biosynthetic protein | VF |
| 12 | ybtU | 3420589..3421749 [+], 1161 | thiazolinyl-S-HMWP1 reductase | VF |
| 13 | ybtT | 3421746..3422549 [+], 804 | yersiniabactin biosynthetic protein | VF |
| 14 | ybtE | 3422553..3424130 [+], 1578 | yersiniabactin siderophore biosynthetic protein | VF |
| 15 | fyuA | 3424261..3426282 [+], 2022 | pesticin/yersiniabactin receptor | VF |
| 16 | KP1_3595 | 3427155..3428081 [+], 927 | conserved hypothetical protein | |
| 17 | alpA2 | 3428221..3428406 [+], 186 | putative transcriptional regulator | |
| 18 | KP1_3597 | 3428595..3429176 [+], 582 | conserved hypothetical protein | |
| 19 | KP1_3599 | 3429618..3429731 [+], 114 | conserved hypothetical protein | |
| 20 | KP1_3600 | 3429775..3430386 [-], 612 | unnamed protein product | TrbB, T4SS component |
| 21 | KP1_3601 | 3430310..3430501 [-], 192 | transposase/IS protein | |
| 22 | KP1_3602 | 3430528..3430620 [-], 93 | putative transposase | |
| 23 | KP1_3603 | 3430596..3431498 [-], 903 | putative transposase | |
| 24 | KP1_3604 | 3431686..3432066 [-], 381 | hypothetical protein | |
| 25 | vagC | 3432509..3432739 [+], 231 | virulence-associated protein | |
| 26 | vagD | 3432736..3433152 [+], 417 | putative DNA binding protein | |
| 27 | KP1_3607 | 3433170..3433433 [+], 264 | hypothetical protein | |
| 28 | iroN | 3433588..3435762 [-], 2175 | enterochelin and dihydrobenzoic acid receptor | VF |
| 29 | iroB | 3436634..3437749 [+], 1116 | putative UDP-glucoronosyl and UDP-glucosyl transferase | VF |
| 30 | iroC | 3437836..3441531 [+], 3696 | putative ABC transporter | VF |
| 31 | iroD | 3441637..3442866 [+], 1230 | ferric enterochelin esterase | VF |
| 32 | pagO | 3443871..3444785 [-], 915 | putative PhoPQ-activated integral membrane protein | |
| 33 | KP1_3616 | 3444858..3445409 [-], 552 | hypothetical protein | |
| 34 | KP1_3617 | 3445596..3445742 [+], 147 | hypothetical protein | |
| 35 | rmpA | 3445784..3446416 [-], 633 | regulator of mucoid phenotype | VF |
| 36 | KP1_3620 | 3446430..3446594 [+], 165 | hypothetical protein | |
| 37 | KP1_3621 | 3446958..3447611 [+], 654 | hypothetical protein | |
| 38 | KP1_3623 | 3447691..3447792 [-], 102 | putative transposase | |
| 39 | KP1_3624 | 3448249..3448434 [-], 186 | transposase/IS protein | TrbB, T4SS component |
| 40 | KP1_3625 | 3448919..3449152 [+], 234 | putative ATP/GTP-binding protein remnant | |
| 41 | virB1 | 3449225..3449935 [+], 711 | type IV secretory pathway VirB1 component | VirB1, T4SS component |
| 42 | virB2 | 3449935..3450228 [+], 294 | type IV secretory pathway VirB2 component | VirB2, T4SS component |
| 43 | virB3-4 | 3450241..3452979 [+], 2739 | type IV secretory pathway VirB3-4 component | VirB4, T4SS component |
| 44 | virB5 | 3452997..3453701 [+], 705 | type IV secretion system VirB5 component | VirB5, T4SS component |
| 45 | KP1_3630 | 3453711..3453938 [+], 228 | conserved hypothetical protein | MagB05, T4SS component |
| 46 | virB6 | 3453950..3455029 [+], 1080 | type IV secretion system VirB6 component | VirB6, T4SS component |
| 47 | virB8 | 3455245..3455928 [+], 684 | type IV secretion system VirB8 component | VirB8, T4SS component |
| 48 | virB9 | 3455925..3456833 [+], 909 | type IV secretory pathway VirB9 component | VirB9, T4SS component |
| 49 | virB10 | 3456877..3458127 [+], 1251 | type IV secretory pathway VirB10 component | VirB10, T4SS component |
| 50 | virB11 | 3458117..3459142 [+], 1026 | type IV secretory pathway VirB11 component | VirB11, T4SS component |
| 51 | KP1_3637 | 3459139..3459537 [+], 399 | hypothetical protein | |
| 52 | KP1_3638 | 3459573..3459875 [+], 303 | conserved hypothetical protein | MagB14, T4SS component |
| 53 | KP1_3639 | 3459918..3460223 [+], 306 | putative dopa decarboxylase protein remnant | |
| 54 | KP1_3640 | 3460334..3460636 [+], 303 | hypothetical protein | |
| 55 | mobB | 3460903..3462792 [+], 1890 | mobilization protein | VirD4, T4SS component |
| 56 | mobC | 3462802..3463548 [+], 747 | mobilization protein | |
| 57 | ardC | 3463744..3464691 [-], 948 | antirestriction protein | |
| 58 | KP1_3644 | 3465676..3466491 [-], 816 | hypothetical protein | |
| 59 | KP1_3645 | 3466792..3467139 [-], 348 | hypothetical protein | |
| 60 | KP1_3646 | 3467139..3468509 [-], 1371 | UBA/THIF-type NAD/FAD binding fold | |
| 61 | KP1_3647 | 3468506..3469633 [-], 1128 | hypothetical protein | |
| 62 | KP1_3648 | 3469728..3470276 [-], 549 | conserved hypothetical protein | |
| 63 | KP1_3649 | 3470319..3471422 [-], 1104 | hypothetical protein | |
| 64 | KP1_3650 | 3471621..3471770 [-], 150 | hypothetical protein | |
| 65 | KP1_3651 | 3472308..3473411 [+], 1104 | hypothetical protein | |
| 66 | KP1_3652 | 3473408..3474379 [-], 972 | hypothetical protein | |
| 67 | KP1_3653 | 3474586..3475563 [-], 978 | putative non-heme chloroperoxidase | |
| 68 | KP1_3654 | 3475615..3476190 [-], 576 | hypothetical protein | |

