The graph information of ICESt2 components from AJ278471 |
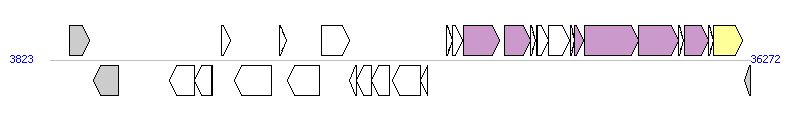 |
| Complete gene list of ICESt2 from AJ278471 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | - | 4728..5657 [+], 930 | putative carboxylate-amine/thiol ligase | |
| 2 | - | 5841..7016 [-], 1176 | putative transposase | |
| 3 | - | 9382..10515 [-], 1134 | putative ATP/GTP-binding protein | |
| 4 | - | 10516..11286 [-], 771 | hypothetical protein | |
| 5 | - | 11316..11339 [-], 24 | ORFV1 protein | |
| 6 | - | 11774..12199 [+], 426 | hypothetical protein | |
| 7 | - | 12382..14103 [-], 1722 | putative ATP/GTP-binding protein | |
| 8 | - | 14462..14767 [+], 306 | hypothetical protein | |
| 9 | sth368IR | 14837..16312 [-], 1476 | R.Sth368I endonuclease | |
| 10 | sth368IM | 16428..17693 [+], 1266 | M.Sth368I methyltransferase | |
| 11 | - | 17725..18024 [-], 300 | hypothetical protein | |
| 12 | arp1 | 18017..18715 [-], 699 | hypothetical transcriptional regulator | |
| 13 | - | 18724..19581 [-], 858 | hypothetical protein | |
| 14 | - | 19679..20995 [-], 1317 | hypothetical protein | |
| 15 | arp2 | 20985..21320 [-], 336 | hypothetical transcriptional regulator | |
| 16 | - | 22183..22479 [+], 297 | hypothetical protein | |
| 17 | - | 22501..22962 [+], 462 | hypothetical protein | OrfM; conjugative transfer; [PMID: 15073287] |
| 18 | - | 22976..24664 [+], 1689 | putative transfer protein | YdcQ, T4SS component |
| 19 | - | 24872..26104 [+], 1233 | putative transfer protein | OrfK; conjugative transfer; [PMID: 15073287] |
| 20 | - | 26119..26349 [+], 231 | hypothetical protein | OrfJ; conjugative transfer; [PMID: 15073287] |
| 21 | - | 26354..26911 [+], 558 | hypothetical protein | |
| 22 | ORFH | 26414..26911 [+], 498 | hypothetical protein | OrfH; conjugative transfer; [PMID: 15073287] |
| 23 | - | 26923..27918 [+], 996 | putative transfer protein | OrfG; conjugative transfer; [PMID: 15073287] |
| 24 | - | 27928..28152 [+], 225 | hypothetical protein | OrfF; conjugative transfer; [PMID: 15073287] |
| 25 | ORFE | 28155..28532 [+], 378 | putative transfer protein | YddD, T4SS component |
| 26 | - | 28578..31082 [+], 2505 | putative ATP/GTP-binding protein | YddE, T4SS component |
| 27 | - | 31094..32974 [+], 1881 | putative transmembrane protein | YddG, T4SS component |
| 28 | - | 32976..33200 [+], 225 | hypothetical protein | |
| 29 | - | 33226..34338 [+], 1113 | putative transfer protein | Orf13_p, T4SS component |
| 30 | xis | 34352..34600 [+], 249 | excisionase | excisionase; [PMID: 15073287] |
| 31 | int | 34600..35946 [+], 1347 | integrase | Integrase |
| 32 | fda | 36030..36272 [-], 243 | putative fructose-1,6-biphosphate aldolase | |