Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 300316 |
| Name | oriT_pSEVA431 |
| Organism | Cloning vector pSEVA431 |
| Sequence Completeness | |
| NCBI accession of oriT (coordinates [strand]) | JX560338 (_) |
| oriT length | 246 nt |
| IRs (inverted repeats) | |
| Location of nic site | |
| Conserved sequence flanking the nic site |
|
| Note | oriT_RP4 |
oriT sequence
Download Length: 246 nt
>oriT_pSEVA431
CTTTTCCGCTGCATAACCCTGCTTCGGGGTCATTATAGCGATTTTTTCGGTATATCCATCCTTTTTCGCACGATATACAGGATTTTGCCAAAGGGTTCGTGTAGACTTTCCTTGGTGTATCCAACGGCGTCAGCCGGGCAGGATAGGTGAAGTAGGCCCACCCGCGAGCGGGTGTTCCTTCTTCACTGTCCCTTATTCGCACCTGGCGGTGCTCAACGGGAATCCTGCTCTGCGAGGCTGGCCGTA
CTTTTCCGCTGCATAACCCTGCTTCGGGGTCATTATAGCGATTTTTTCGGTATATCCATCCTTTTTCGCACGATATACAGGATTTTGCCAAAGGGTTCGTGTAGACTTTCCTTGGTGTATCCAACGGCGTCAGCCGGGCAGGATAGGTGAAGTAGGCCCACCCGCGAGCGGGTGTTCCTTCTTCACTGTCCCTTATTCGCACCTGGCGGTGCTCAACGGGAATCCTGCTCTGCGAGGCTGGCCGTA
Visualization of oriT structure (The oriT was characterized experimentally)
oriT secondary structure
Predicted by RNAfold.
Download structure file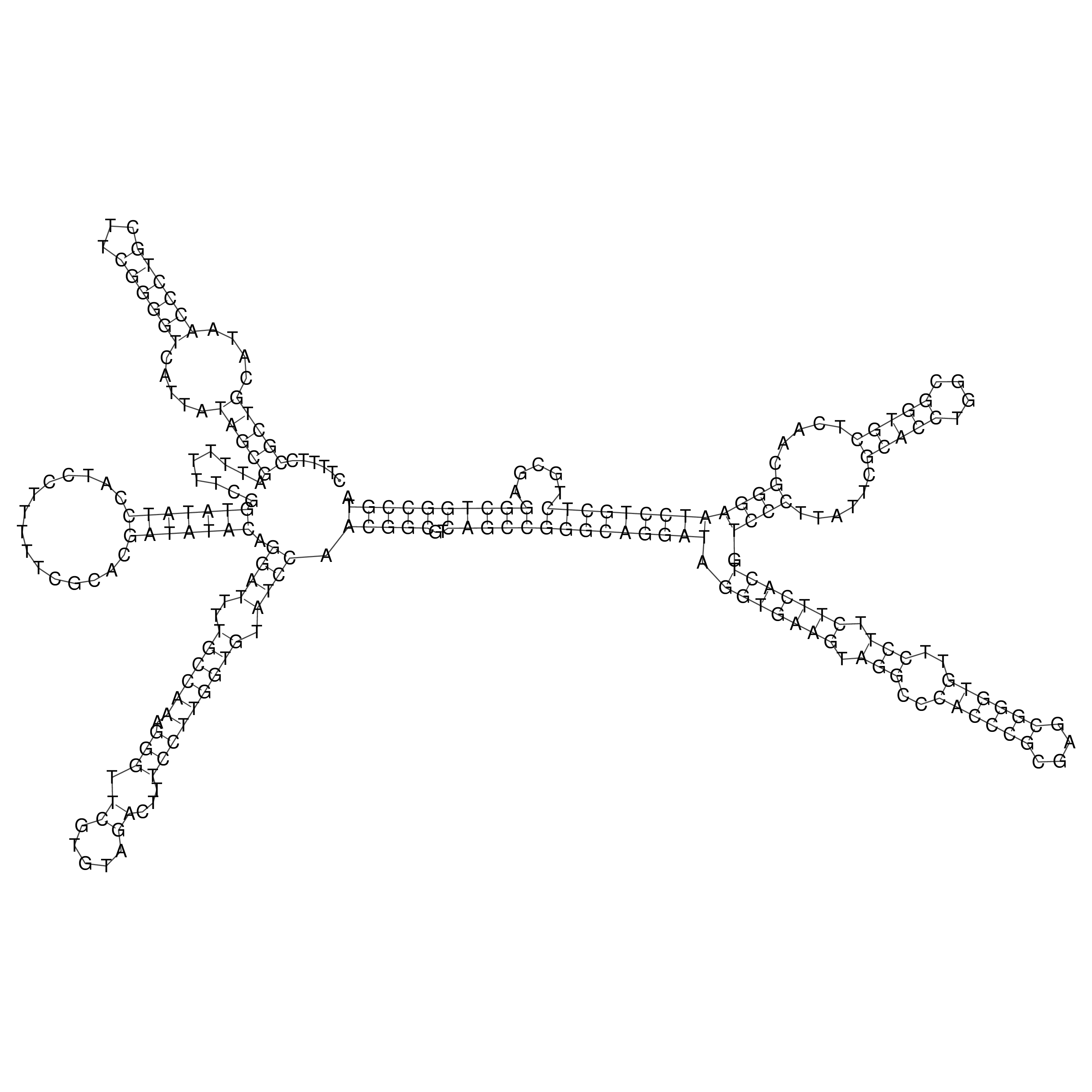
Reference
[1] Silva-Rocha R et al. (2013) The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res. 41(Database issue):D666-75. [PMID:23180763]
Host bacterium
| ID | 529 | GenBank | JX560338 |
| Plasmid name | Cloning vector pSEVA431 | Incompatibility group | - |
| Plasmid size | 3182 bp | Coordinate of oriT [Strand] | |
| Host baterium | Cloning vector pSEVA431 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |