Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200560 |
| Name | oriT_IME_LspYL32_NS_1 |
| Organism | Lachnoclostridium sp. YL32 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | CP015399 (136..170 [+], 35 nt) |
| oriT length | 35 nt |
| IRs (inverted repeats) | 5..11, 15..21 (ACTTTGC..GCAAAGT) |
| Location of nic site | 29..30 |
| Conserved sequence flanking the nic site |
GTGTGTTATA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 35 nt
>oriT_IME_LspYL32_NS_1
CACGACTTTGCGGAGCAAAGTGTAGTGTGTTATAC
CACGACTTTGCGGAGCAAAGTGTAGTGTGTTATAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file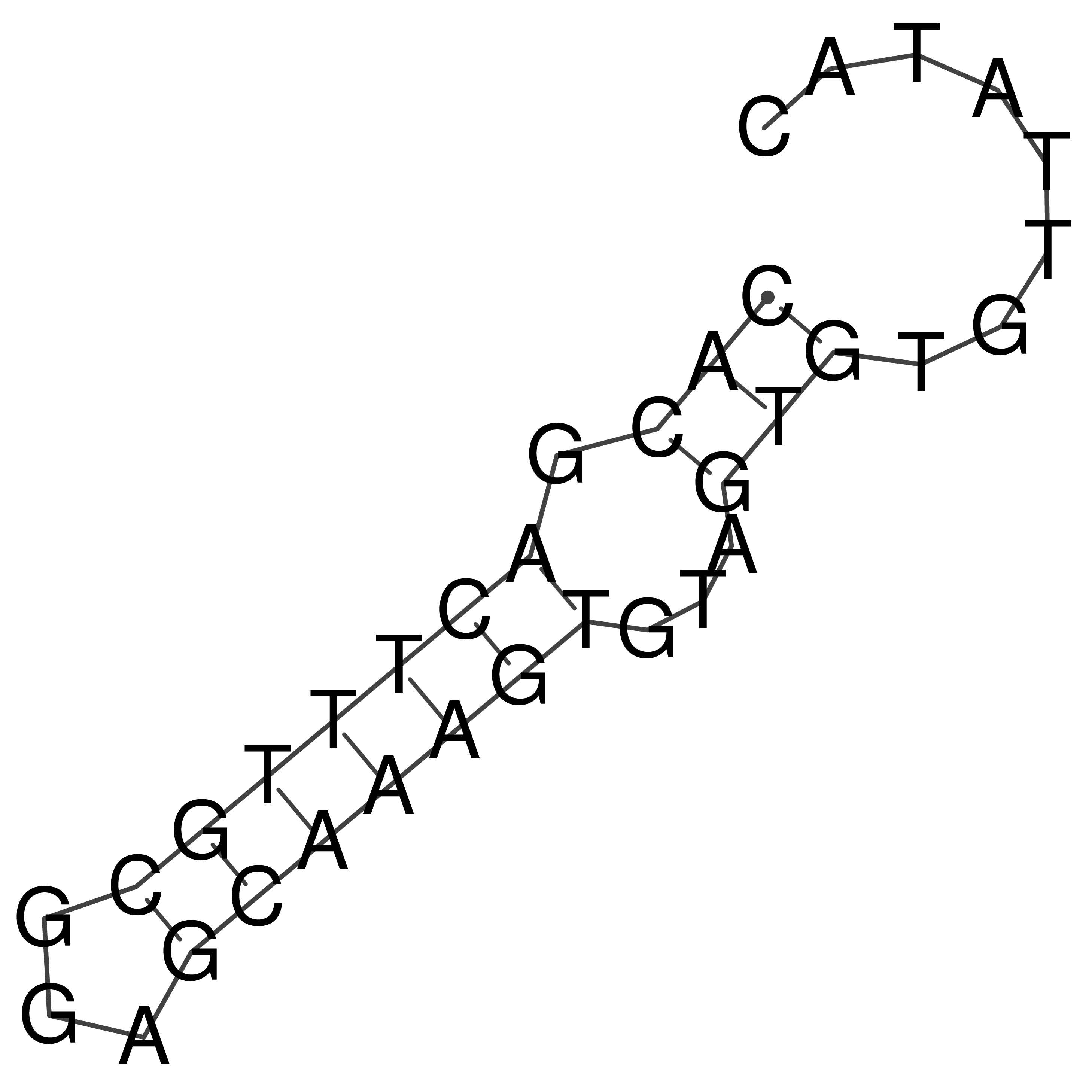
Relaxase
| ID | 14589 | GenBank | ANU46859 |
| Name | Mob_Pre_A4V08_14585_IME_LspYL32_NS_1 |
UniProt ID | _ |
| Length | 314 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 314 a.a. Molecular weight: 36383.80 Da Isoelectric Point: 10.3227
>ANU46859.1 plasmid recombination enzyme [Lachnoclostridium sp. YL32]
MAQHAILRFEKHKGHPAGPLEAHHERKKEQYASNPDIDTSRSKYNFHIVKPDGRYYHFIQSRIEQARCRT
RKDSTRFVDTLITASPEFFKGKSPKEIAAYFQRAADFLIDRVGRENIVSAMVHMDEKTPHLHLVFVPLTK
DNRLCAKEIIGNRANLTKWQDDFHACMVEQYPDLERGESASKTGRKHIPTRLFKQAVNLSKQARAIEAVL
SGITPLNAGKKKEEALSMLKKWFPQMENFSGQLKKYKVTINDLLAENEKLEVRAKASEKGKMNDTMERAK
LKSELDDMRRLVDRIPPEILAELKRQQRQHGKER
MAQHAILRFEKHKGHPAGPLEAHHERKKEQYASNPDIDTSRSKYNFHIVKPDGRYYHFIQSRIEQARCRT
RKDSTRFVDTLITASPEFFKGKSPKEIAAYFQRAADFLIDRVGRENIVSAMVHMDEKTPHLHLVFVPLTK
DNRLCAKEIIGNRANLTKWQDDFHACMVEQYPDLERGESASKTGRKHIPTRLFKQAVNLSKQARAIEAVL
SGITPLNAGKKKEEALSMLKKWFPQMENFSGQLKKYKVTINDLLAENEKLEVRAKASEKGKMNDTMERAK
LKSELDDMRRLVDRIPPEILAELKRQQRQHGKER
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 499 | Element type | IME (Integrative mobilizable element) |
| Element name | IME_LspYL32_NS_1 | GenBank | CP015399 |
| Element size | 7225339 bp | Coordinate of oriT [Strand] | 136..170 [+] |
| Host bacterium | Lachnoclostridium sp. YL32 | Coordinate of element | 3171611..3179237 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21 |