Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200549 |
| Name | oriT_SGI1-B2 |
| Organism | Proteus mirabilis strain PmSC17 Salmonella |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | KP116299 (30640..30763 [+], 124 nt) |
| oriT length | 124 nt |
| IRs (inverted repeats) | 65..70, 76..81 (CGGGGG..CCCCCG) 43..49, 51..57 (CCTTGAG..CTCAAGG) 8..14, 19..25 (CGCGCAC..GTGCGCG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 124 nt
>oriT_SGI1-B2
TATAATTCGCGCACATTCGTGCGCGGTGCGAAAGCCTAGAGCCCTTGAGGCTCAAGGCTTCCGTCGGGGGCTCTACCCCCGTCTCTGTTTACGCCTACGGCGACAGAGACGGGGTGGAGCATAG
TATAATTCGCGCACATTCGTGCGCGGTGCGAAAGCCTAGAGCCCTTGAGGCTCAAGGCTTCCGTCGGGGGCTCTACCCCCGTCTCTGTTTACGCCTACGGCGACAGAGACGGGGTGGAGCATAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file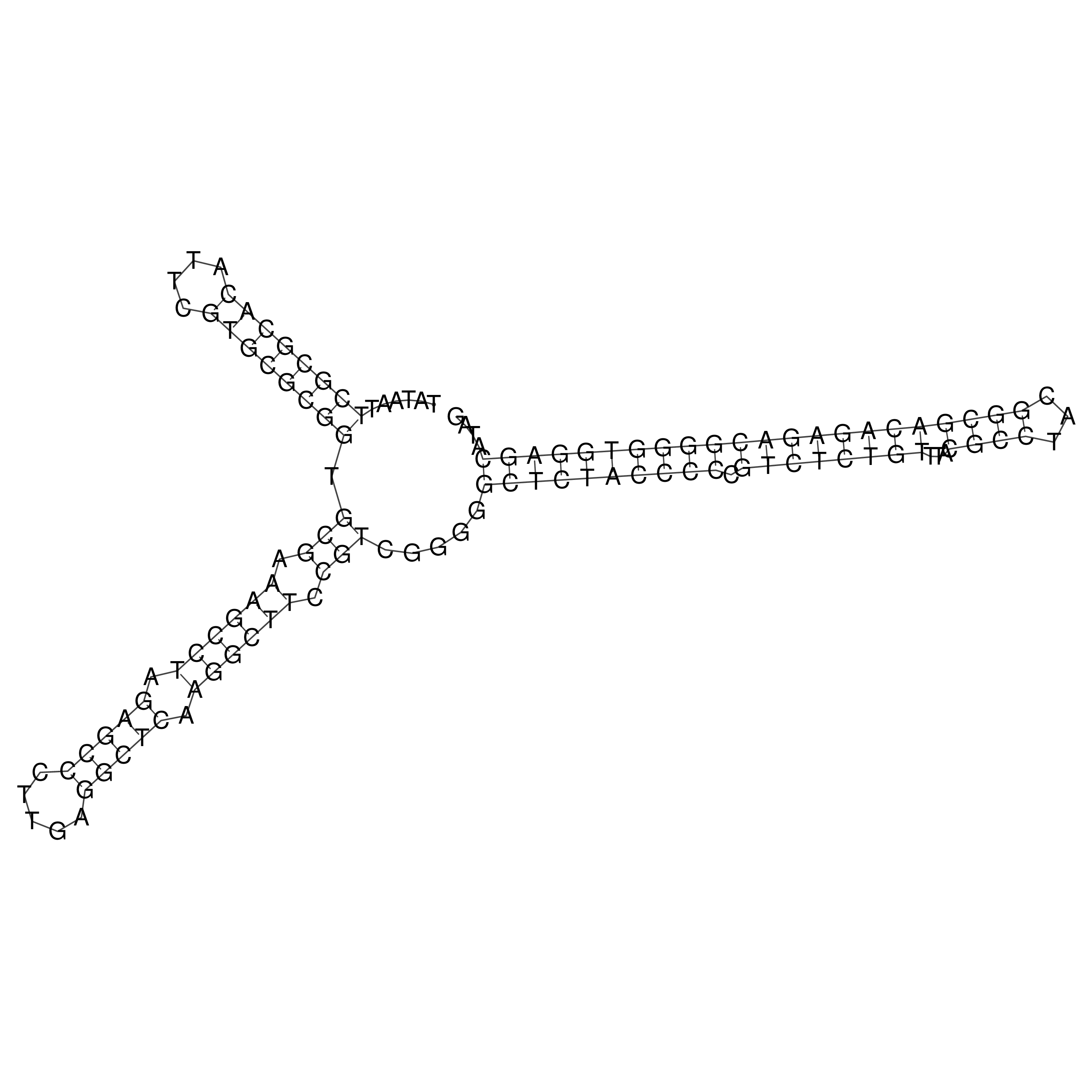
Relaxase
| ID | 14584 | GenBank | AJS16039 |
| Name | Replic_Relax_-_SGI1-B2 |
UniProt ID | _ |
| Length | 101 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 101 a.a. Molecular weight: 11477.11 Da Isoelectric Point: 7.5204
>AJS16039.1 PadR family transcriptional regulator [Proteus mirabilis]
MTDKDLYGGLIRLHILHHAAEEPVFGLGIIEELRRHGYEMSAGTVYPMLHGLEKKGYLTSRHERTGRRER
RVYDITEQGRTALADAKTKVKELFGELVEGG
MTDKDLYGGLIRLHILHHAAEEPVFGLGIIEELRRHGYEMSAGTVYPMLHGLEKKGYLTSRHERTGRRER
RVYDITEQGRTALADAKTKVKELFGELVEGG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 491 | Element type | IME (Integrative mobilizable element) |
| Element name | SGI1-B2 | GenBank | KP116299 |
| Element size | 48650 bp | Coordinate of oriT [Strand] | 30640..30763 [+] |
| Host bacterium | Proteus mirabilis strain PmSC17 Salmonella | Coordinate of element | 139..46067 |
Cargo genes
| Drug resistance gene | dfrA17, aadA5, qacE, sul1, mph(A), blaCARB-2 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |