Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200513 |
| Name | oriT_TnGBS2.3 |
| Organism | Streptococcus agalactiae strain Wc3 DDE transposase UPF0236 family (TnGBS2.3_01) hypothetical protein (TnGBS2.3_02) |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | KC492040 (4484..4516 [-], 33 nt) |
| oriT length | 33 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 33 nt
>oriT_TnGBS2.3
GCATGTGTCCGGACACGTGCTAATCTTGCCAGA
GCATGTGTCCGGACACGTGCTAATCTTGCCAGA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file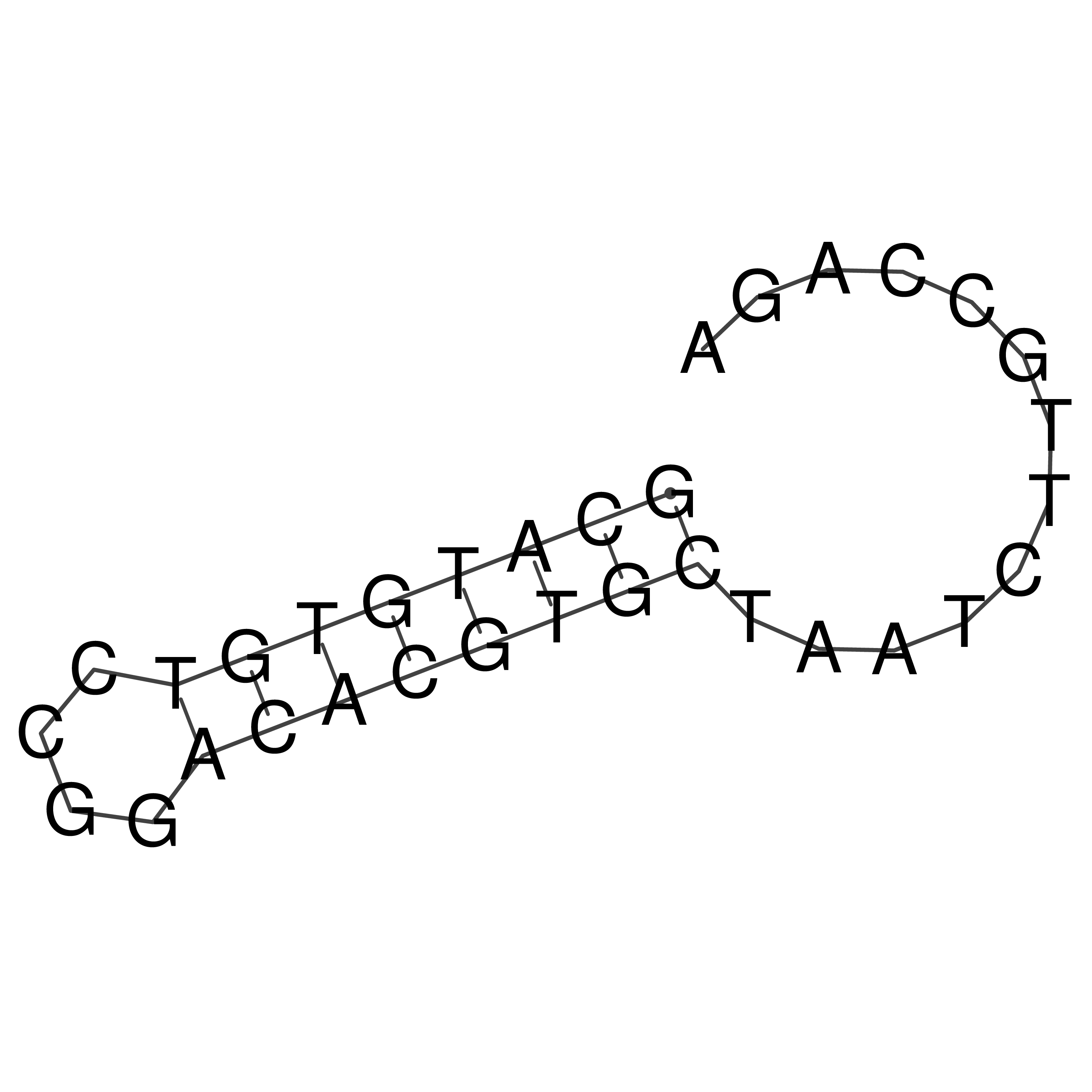
Relaxase
| ID | 14569 | GenBank | AGO89368 |
| Name | Relaxase_TnGBS2.3_03_TnGBS2.3 |
UniProt ID | _ |
| Length | 546 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 546 a.a. Molecular weight: 63831.43 Da Isoelectric Point: 7.2330
>AGO89368.1 putative relaxase [Streptococcus agalactiae]
MVVTKVFQIKQIKNLKRAIDYITRDDATLIKVEQHDEDDAFDYTLTADGEVHKKLISGHHLIDPGDRDAI
YDDFILLKQSVDELYDNATLSDLKNDNRVLAHHIVQSFSPDDKLTPEEVNEIGRKTALEFTGGDYQFVVA
THVDKGFLHNHIIFNTTNEVTLKKFRWQKNTARNLFQISNKHAELYGAKILEPKLRTSYTDYSAWRRQNN
FSYEIKQRLNFLLKHSISIEDFQQKAKALDLHIDFSGKFAKYRLLVPLDGKLQEKNTRDRTLSKKRIYSF
EQIQKRVAMNEVVYTVDHIKEKYEEEEAKKAEDFEMKVRIEPWQISEITRQSIHLPVTFGLEQKGTVSIP
ARLLDQNEDGSFTAYFKKKDYFYFINPDKSTDNRFIQGSTLVKQLSLNSGEVILTKNWEMTELERLVKEF
NFLSANKVTNSKQFQEMQERFLEQLEKTDQTLAQLDSKVAYLNKVLGALSDYQDGLVPSEVTMAILEKAK
VAKNTDRNLIRKEIKELQIEREALQEARDRIVKDYDFSHKLTQEYSPKESRKKSII
MVVTKVFQIKQIKNLKRAIDYITRDDATLIKVEQHDEDDAFDYTLTADGEVHKKLISGHHLIDPGDRDAI
YDDFILLKQSVDELYDNATLSDLKNDNRVLAHHIVQSFSPDDKLTPEEVNEIGRKTALEFTGGDYQFVVA
THVDKGFLHNHIIFNTTNEVTLKKFRWQKNTARNLFQISNKHAELYGAKILEPKLRTSYTDYSAWRRQNN
FSYEIKQRLNFLLKHSISIEDFQQKAKALDLHIDFSGKFAKYRLLVPLDGKLQEKNTRDRTLSKKRIYSF
EQIQKRVAMNEVVYTVDHIKEKYEEEEAKKAEDFEMKVRIEPWQISEITRQSIHLPVTFGLEQKGTVSIP
ARLLDQNEDGSFTAYFKKKDYFYFINPDKSTDNRFIQGSTLVKQLSLNSGEVILTKNWEMTELERLVKEF
NFLSANKVTNSKQFQEMQERFLEQLEKTDQTLAQLDSKVAYLNKVLGALSDYQDGLVPSEVTMAILEKAK
VAKNTDRNLIRKEIKELQIEREALQEARDRIVKDYDFSHKLTQEYSPKESRKKSII
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 17186 | GenBank | AGO89374 |
| Name | t4cp2_TnGBS2.3_09_TnGBS2.3 |
UniProt ID | _ |
| Length | 681 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 681 a.a. Molecular weight: 78336.99 Da Isoelectric Point: 5.0886
>AGO89374.1 VirD4 protein domain protein [Streptococcus agalactiae]
MELKRKRLLPYLLLSLLLAYLTHRAYVLYCMAPQADMTNLFAPYTYVFEKITEPPYFYWNTSPLAILSAL
VGFFVGLLFYLKIRLDGNYRHGEESGSACFATAQELKGFQDEESRNNMIFSREAQMGLFNKRLPFERQLN
KNVLVAGLPGDGKTYTYVKPNLMQMNSSFIVTDPKGLLVREVGTMLKENGYQIKIFDLVNLTNSDMFNPF
RYMRSELDIDRITEAIVEGTKKGDREGEDFWNQAKLLLNRALLGYLYFDSQVRHYEPNLSMVADLLRNLK
RPNEKELSAVEKMFKQLEEKLPGNYACRQWELFNSNFEAETRTGVLSIVATQYSIFDHDAVTNLIKTDTM
EMDTWNTEKTAVFVAISETNKAYSFLASTFFTVVFDQLTHSADAIIQGEKKGYELKDLLHVQFYFDEFAN
CGKIPHFNEVLASIRSREMSIKIIIQAISQLDSLYGVPARKSMVNNCATLLFLGTNDEDTMKYFSMRAGK
QTIIQTSYSEQRGHRISGTTSRQAHQRDLMTPDEIARIGVDEALVFISKQNVFRDKKAMVSNHPMKDRLA
NHYQDKTWYHYKRFMEEGAEFIDAVMTGQIPEDRIWAPDMSDYQAFLEENGFDNTRQSAPETSVHNQVPV
TNPEMPSESPSQTPETSTKLVDMDTGEIFELPPEDREGFGEVSFDDQRVEI
MELKRKRLLPYLLLSLLLAYLTHRAYVLYCMAPQADMTNLFAPYTYVFEKITEPPYFYWNTSPLAILSAL
VGFFVGLLFYLKIRLDGNYRHGEESGSACFATAQELKGFQDEESRNNMIFSREAQMGLFNKRLPFERQLN
KNVLVAGLPGDGKTYTYVKPNLMQMNSSFIVTDPKGLLVREVGTMLKENGYQIKIFDLVNLTNSDMFNPF
RYMRSELDIDRITEAIVEGTKKGDREGEDFWNQAKLLLNRALLGYLYFDSQVRHYEPNLSMVADLLRNLK
RPNEKELSAVEKMFKQLEEKLPGNYACRQWELFNSNFEAETRTGVLSIVATQYSIFDHDAVTNLIKTDTM
EMDTWNTEKTAVFVAISETNKAYSFLASTFFTVVFDQLTHSADAIIQGEKKGYELKDLLHVQFYFDEFAN
CGKIPHFNEVLASIRSREMSIKIIIQAISQLDSLYGVPARKSMVNNCATLLFLGTNDEDTMKYFSMRAGK
QTIIQTSYSEQRGHRISGTTSRQAHQRDLMTPDEIARIGVDEALVFISKQNVFRDKKAMVSNHPMKDRLA
NHYQDKTWYHYKRFMEEGAEFIDAVMTGQIPEDRIWAPDMSDYQAFLEENGFDNTRQSAPETSVHNQVPV
TNPEMPSESPSQTPETSTKLVDMDTGEIFELPPEDREGFGEVSFDDQRVEI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 12141..27172
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| TnGBS2.3_08 | 9689..10054 | - | 366 | AGO89373 | hypothetical protein | - |
| TnGBS2.3_09 | 10096..12141 | - | 2046 | AGO89374 | VirD4 protein domain protein | - |
| TnGBS2.3_10 | 12141..12632 | - | 492 | AGO89375 | hypothetical protein | cd411 |
| TnGBS2.3_11 | 12654..13439 | - | 786 | AGO89376 | hypothetical protein | - |
| TnGBS2.3_12 | 13731..14336 | - | 606 | AGO89377 | hypothetical protein | prgL |
| TnGBS2.3_13 | 14357..16675 | - | 2319 | AGO89378 | VirB1 domain protein | prgK |
| TnGBS2.3_14 | 17082..17408 | - | 327 | AGO89379 | hypothetical protein | - |
| TnGBS2.3_15 | 17432..18052 | - | 621 | AGO89380 | hypothetical protein | - |
| TnGBS2.3_16 | 18018..20417 | - | 2400 | AGO89381 | VirB4type IV secretion/conjugal transfer ATPase, VirB4 family protein | virb4 |
| TnGBS2.3_17 | 20359..20736 | - | 378 | AGO89382 | hypothetical protein | - |
| TnGBS2.3_18 | 20780..21238 | - | 459 | AGO89383 | hypothetical protein | - |
| TnGBS2.3_19 | 21231..21527 | - | 297 | AGO89384 | hypothetical protein | - |
| TnGBS2.3_20 | 21564..22316 | - | 753 | AGO89385 | VirB6 domain protein | prgHb |
| TnGBS2.3_21 | 22367..22948 | - | 582 | AGO89386 | hypothetical protein | - |
| TnGBS2.3_22 | 23575..26397 | - | 2823 | AGO89387 | LPXTG protein | prgB |
| TnGBS2.3_23 | 26450..27172 | - | 723 | AGO89388 | LPXTG protein | prgC |
| TnGBS2.3_24 | 27192..29453 | - | 2262 | AGO89389 | LPXTG protein | - |
| TnGBS2.3_25 | 29470..29769 | - | 300 | AGO89390 | hypothetical protein | - |
| TnGBS2.3_26 | 30533..31603 | - | 1071 | AGO89391 | RepA Nterminal domain protein | - |
Host bacterium
| ID | 456 | Element type | ICE (Integrative and conjugative element) |
| Element name | TnGBS2.3 | GenBank | KC492040 |
| Element size | 34350 bp | Coordinate of oriT [Strand] | 4484..4516 [-] |
| Host bacterium | Streptococcus agalactiae strain Wc3 DDE transposase UPF0236 family (TnGBS2.3_01) hypothetical protein (TnGBS2.3_02) | Coordinate of element | 1347..33853 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA7, AcrIIA21 |