Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200511 |
| Name | oriT_Tn6198 |
| Organism | Listeria monocytogenes strain TTH-2007 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | JX120102 (5792..5924 [+], 133 nt) |
| oriT length | 133 nt |
| IRs (inverted repeats) | 65..70, 83..88 (AAATCC..GGATTT) 6..12, 23..29 (ACCCCCC..GGGGGGT) |
| Location of nic site | 75..76 |
| Conserved sequence flanking the nic site |
TTTGGTTACA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 133 nt
>oriT_Tn6198
ACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATAC
ACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file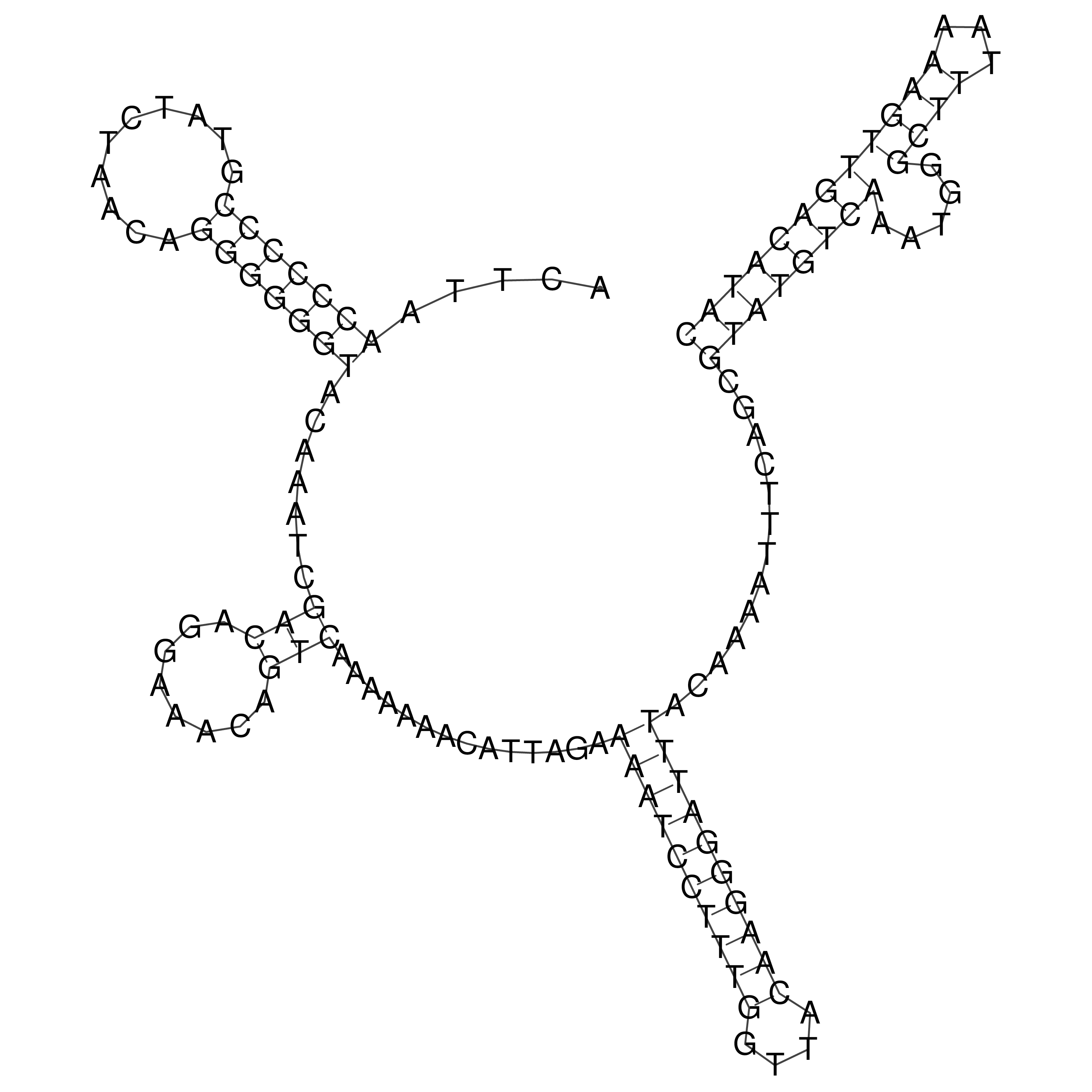
Relaxase
| ID | 14565 | GenBank | AGC70174 |
| Name | Rep_trans_-_Tn6198 |
UniProt ID | _ |
| Length | 329 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 329 a.a. Molecular weight: 39099.72 Da Isoelectric Point: 7.0754
>AGC70174.1 hypothetical protein [Listeria monocytogenes]
MLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGRGC
RQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRSGE
LVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLLVY
DNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQVAP
TLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
MLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGRGC
RQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRSGE
LVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLLVY
DNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQVAP
TLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 7935 | GenBank | AGC70191 |
| Name | AGC70191_Tn6198 |
UniProt ID | _ |
| Length | 405 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 405 a.a. Molecular weight: 47021.92 Da Isoelectric Point: 9.7978
>AGC70191.1 integrase [Listeria monocytogenes]
MSEKRRDNKGRILKTGESQRKDGRYLYKYIDSFGEPQFVYSWKLVATDRVPAGKRDCISLREKIAELQKD
IHDGIDVVGKKMTLCQLYAKQNAQRPKVRKNTETGRKYLMDILKKDKLGVRSIDSIKPSDAKEWAIRMSE
NGYAYQTINNYKRSLKASFYIAIQDDCVRKNPFDFQLKAVLDDDTVPKTVLTEEQEEKLLAFAKADKTYS
KNYDEILILLKTGLRISEFGGLTLPDLDFENRLVNIDHQLLRDTEIGYYIETPKTKSGERQVPMVEEAYQ
AFKRVLANRKNDKRVEIDGYSDFLFLNRKNYPKVASDYNGMMKGLVKKYNKYNEDKLPHITPHSLRHTFC
TNYANAGMNPKALQYIMGHANIAMTLNYYAHATFDSAMAEMKRLNKEKQQERLVA
MSEKRRDNKGRILKTGESQRKDGRYLYKYIDSFGEPQFVYSWKLVATDRVPAGKRDCISLREKIAELQKD
IHDGIDVVGKKMTLCQLYAKQNAQRPKVRKNTETGRKYLMDILKKDKLGVRSIDSIKPSDAKEWAIRMSE
NGYAYQTINNYKRSLKASFYIAIQDDCVRKNPFDFQLKAVLDDDTVPKTVLTEEQEEKLLAFAKADKTYS
KNYDEILILLKTGLRISEFGGLTLPDLDFENRLVNIDHQLLRDTEIGYYIETPKTKSGERQVPMVEEAYQ
AFKRVLANRKNDKRVEIDGYSDFLFLNRKNYPKVASDYNGMMKGLVKKYNKYNEDKLPHITPHSLRHTFC
TNYANAGMNPKALQYIMGHANIAMTLNYYAHATFDSAMAEMKRLNKEKQQERLVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 17182 | GenBank | AGC70173 |
| Name | tcpA_-_Tn6198 |
UniProt ID | _ |
| Length | 461 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 461 a.a. Molecular weight: 53370.27 Da Isoelectric Point: 9.0687
>AGC70173.1 hypothetical protein [Listeria monocytogenes]
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKN
GLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVEAKDGKLRL
MKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLL
SCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLG
RQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSV
ISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKN
GLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVEAKDGKLRL
MKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLL
SCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLG
RQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSV
ISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 3627..15036
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| Locus_0 | 200..319 | + | 120 | AGC70167 | hypothetical protein | - |
| Locus_1 | 578..814 | + | 237 | AGC70168 | hypothetical protein | - |
| Locus_2 | 821..2773 | + | 1953 | AGC70169 | hypothetical protein | - |
| Locus_3 | 2845..3342 | - | 498 | AGC70170 | dihydrofolate reductase | - |
| Locus_4 | 3627..3941 | + | 315 | AGC70171 | hypothetical protein | orf23 |
| Locus_5 | 3957..4343 | + | 387 | AGC70172 | hypothetical protein | orf23 |
| Locus_6 | 4372..5757 | + | 1386 | AGC70173 | hypothetical protein | virb4 |
| Locus_7 | 6151..7140 | + | 990 | AGC70174 | hypothetical protein | - |
| Locus_8 | 7183..7404 | + | 222 | AGC70175 | hypothetical protein | orf19 |
| Locus_9 | 7521..8018 | + | 498 | AGC70176 | hypothetical protein | - |
| Locus_10 | 7993..8499 | + | 507 | AGC70177 | hypothetical protein | orf17a |
| Locus_11 | 8483..10930 | + | 2448 | AGC70178 | hypothetical protein | virb4 |
| Locus_12 | 10933..13197 | + | 2265 | AGC70179 | hypothetical protein | orf15 |
| Locus_13 | 13106..14107 | + | 1002 | AGC70180 | hypothetical protein | orf14 |
| Locus_14 | 14104..15036 | + | 933 | AGC70181 | hypothetical protein | orf13 |
| Locus_15 | 15311..15397 | + | 87 | AGC70182 | tet(M) leader peptide | - |
| Locus_16 | 15413..17332 | + | 1920 | AGC70183 | tetracycline resistance | - |
| Locus_17 | 17430..17618 | + | 189 | AGC70184 | hypothetical protein | - |
| Locus_18 | 17678..18031 | - | 354 | AGC70185 | hypothetical protein | - |
| Locus_19 | 18236..18307 | + | 72 | AGC70186 | hypothetical protein | - |
| Locus_20 | 18485..18958 | + | 474 | AGC70187 | hypothetical protein | - |
| Locus_21 | 18955..19185 | + | 231 | AGC70188 | hypothetical protein | - |
| Locus_22 | 19411..19662 | - | 252 | AGC70189 | hypothetical protein | - |
| Locus_23 | 19646..19849 | + | 204 | AGC70190 | excisionase | - |
Host bacterium
| ID | 454 | Element type | ICE (Integrative and conjugative element) |
| Element name | Tn6198 | GenBank | JX120102 |
| Element size | 21322 bp | Coordinate of oriT [Strand] | 5792..5924 [+] |
| Host bacterium | Listeria monocytogenes strain TTH-2007 | Coordinate of element | 1..21322 |
Cargo genes
| Drug resistance gene | dfrG, tet(M) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |