Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200503 |
| Name | oriT_ICESpnDCC1738 |
| Organism | Streptococcus pneumoniae integrative and |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | HG799492 (43792..43924 [-], 133 nt) |
| oriT length | 133 nt |
| IRs (inverted repeats) | 65..70, 83..88 (AAATCC..GGATTT) 6..12, 23..29 (ACCCCCC..GGGGGGT) |
| Location of nic site | 75..76 |
| Conserved sequence flanking the nic site |
TTTGGTTACA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 133 nt
ACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file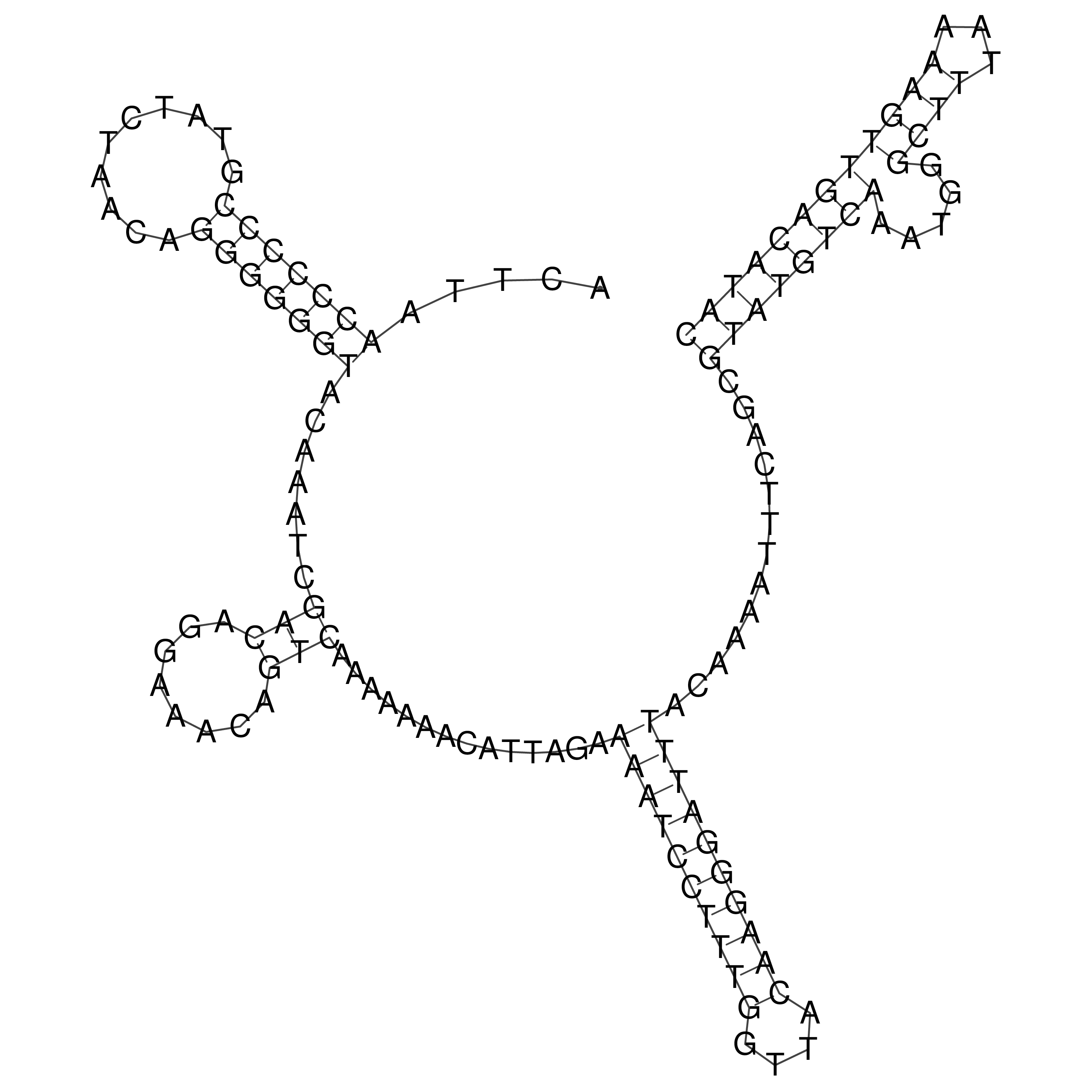
Relaxase
| ID | 14550 | GenBank | CDL73806 |
| Name | Rep_trans_-_ICESpnDCC1738 |
UniProt ID | _ |
| Length | 401 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 401 a.a. Molecular weight: 47398.10 Da Isoelectric Point: 6.5091
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 14551 | GenBank | CDL73829 |
| Name | Relaxase_-_ICESpnDCC1738 |
UniProt ID | _ |
| Length | 609 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 609 a.a. Molecular weight: 73453.95 Da Isoelectric Point: 9.2584
MVITKHFAIHGKSYRRKIIKYILNPDKTKNLALVSDYGMRNFLDFPSYDEMVQMYHENFISNDTLYNFRH
ARLEEKQRKIHAHHIIQSFSPDDHLTPEQINRIGYETAKELTGGRFRFIVATHVDKDHIHNHIILNSIDK
NSDKKFLWDYKAERNLRMVSDRLSKIVGAKIIENRYSHHQYEVYRKTNYKYEIKQRVYFLIENSKNFEDF
KKKAKDLHLKIDFRHKHVTFFMTDSNMKQVVRDNKLNRKQPYNETYFKKKFVQREIINILEFLLPKMKNM
NELIQQAEFFGLKIILKAKHVLFEFDGIKFSEQELVKSNQYSVSYFQDYFNNKNDTFGLDNKNLVELYNE
EKLIKEKKLPTEDMVWKSYQDFKRNRDAVHEFEVELNLNQIEEVVDDGIYIKVQFGIRQEGLIFVPNIQI
NMEEEKVKVFLRETSSYYVYHKDLAEKNRFMKGKTLIRQFNLQYEQQYMYRRISLSKIKEKIEQLDFLMS
AENSPNDFEDITNDFIAQISYLENMIEQVQNKIDDLTNLEEVLLNNTTNSSSNLENSIQDKSSVDKIEKD
LYIYKGKIEKLKEQHREAINLFEMFNKTIKKYKKKQNMKSIEENEIHLE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 7925 | GenBank | CDL73794 |
| Name | CDL73794_ICESpnDCC1738 |
UniProt ID | _ |
| Length | 361 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 361 a.a. Molecular weight: 41696.88 Da Isoelectric Point: 9.6071
MATDRVPAGKRDCISLREKIAELQKDIHDGIDVVGKKMTLCQLYAKQNAQRPKVRKNTETGRKYLMDILK
KDKLGVRSIDSIKPSDAKEWAIRMSENGYAYQTINNYKRSLKASFYIAIQDDCVRKNPFDFQLKAVLDDD
TVPKTVLTEEQEEKLLAFAKADKTYSKNYDEILILLKTGLRISEFGGLTLPDLDFENRLVNIDHQLLRDT
EIGYYIETPKTKSGERQVPMVEEAYQAFKRVLANRKNDKRVEIDGYSDFLFLNRKNYPKVASDYNGMMKG
LVKKYNKYNEDKLPHITPHSLRHTFCTNYANAGMNPKALQYIMGHANIAMTLNYYAHATFDSAMAEMKRL
NKEKQQERLVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 17167 | GenBank | CDL73780 |
| Name | t4cp2_-_ICESpnDCC1738 |
UniProt ID | _ |
| Length | 625 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 625 a.a. Molecular weight: 72374.70 Da Isoelectric Point: 9.6194
MYSGKKFLLFSLLGILLGYLFHRLTLLYDSYTGNTLDKWTHLLMEGQDEVLQSPWNVSFTGKSSAFFLLG
FVMMLLVYLYLETGKKQYREGVEYGSARFGTLKEKKLFYGKEFSHDTILAQDVRLTLLDKKPPQYDRNKN
IAVIGGSGSGKTFRFVKPNLIQMNSSNIVVDPKDHLAEKTGKLFLERGYQVKVLDLVNMKNSDGFNPFRY
IETENDLNRMLTVYFNNTKGSGSRSDPFWDEASMTLVRALASYLVDFYNPPKTREQLIEESRLSQKEYQN
LLKRQKKEVEERKKRGRYPSFAEISKLIKHLSKGENQEKSVLEILFENYAKKYGTENFTMRNWADFQNYK
DKTLDSVIAVTTAKFALFNIQSVMDLTKRDTLDMKTWGKEKSMVYLVIPDNDSTFRFLSALFFSTVFQTL
TRQADIDFKGQLPLHVRVYLDEFANIGEIPDFAEQTSTVRSRNMSLVPILQNIAQLQGLYKEKEAWKTIL
GNCDSLVYLGGNDEDTFKFMSGLLGKQTIDVRNTSRSFGQTGSGSLSHQKIARDLMTPDEVGNMKRHECL
VRIANMPVFKSKKYNSTKHPNWKYLANQENDERWWNYQINPLNQSQENHLEGLRIRDLTFESSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 17168 | GenBank | CDL73807 |
| Name | tcpA_-_ICESpnDCC1738 |
UniProt ID | _ |
| Length | 460 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 460 a.a. Molecular weight: 53241.16 Da Isoelectric Point: 9.1761
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKKITYFPKMYYRLKNG
LIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVEAKDGKLRLM
KNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLLS
CIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLGR
QAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSVI
SEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 2391..17619
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| Locus_0 | 1..174 | + | 174 | CDL73766 | conserved hypothetical protein | - |
| Locus_1 | 171..950 | + | 780 | CDL73767 | putative replication initiator protein | - |
| Locus_2 | 1049..2407 | + | 1359 | CDL73768 | methyl transferase | - |
| Locus_3 | 1080..1622 | + | 543 | CDL73769 | methyl transferase | - |
| Locus_4 | 2391..2840 | + | 450 | CDL73770 | conserved hypothetical protein | gbs1369 |
| Locus_5 | 2833..3213 | + | 381 | CDL73771 | conserved hypothetical protein | - |
| Locus_6 | 3226..3459 | + | 234 | Protein_6 | putative uncharacterized protein | - |
| Locus_7 | 3534..4049 | + | 516 | CDL73773 | Putative membrane protein | - |
| Locus_8 | 4341..4976 | + | 636 | CDL73774 | hypothetical protein | - |
| Locus_9 | 5031..5273 | + | 243 | CDL73775 | caax amino protease family | - |
| Locus_10 | 5401..5991 | + | 591 | CDL73776 | conserved hypothetical protein | - |
| Locus_11 | 5991..6827 | + | 837 | CDL73777 | abortive infection protein AbiGII, putative | - |
| Locus_12 | 7109..7408 | + | 300 | CDL73778 | conserved hypothetical protein | - |
| Locus_13 | 7422..7886 | + | 465 | CDL73779 | conserved hypothetical protein | gbs1365 |
| Locus_14 | 7886..9763 | + | 1878 | CDL73780 | putative conjugal transfer protein TraG | virb4 |
| Locus_15 | 9784..10026 | + | 243 | CDL73781 | conserved hypothetical protein | prgF |
| Locus_16 | 10043..10897 | + | 855 | CDL73782 | membrane protein, putative | prgHb |
| Locus_17 | 10951..11310 | + | 360 | CDL73783 | conserved hypothetical protein | prgIc |
| Locus_18 | 11261..13618 | + | 2358 | CDL73784 | putative conjugal transfer protein | virb4 |
| Locus_19 | 13630..16443 | + | 2814 | CDL73785 | putative conjugal transfer protein | prgK |
| Locus_20 | 16745..16858 | - | 114 | CDL73786 | Conserved hypothetical protein | - |
| Locus_21 | 17209..17619 | + | 411 | CDL73787 | Hypothetical protein | cd424 |
Region 2: 34676..51527
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| Locus_29 | 29863..30066 | - | 204 | CDL73795 | excisionase | - |
| Locus_30 | 30754..31176 | - | 423 | CDL73796 | putative conjugative transposon regulatory protein | - |
| Locus_31 | 31681..32034 | + | 354 | CDL73797 | putative conjugative transposon regulatory protein | - |
| Locus_32 | 32380..34314 | - | 1935 | CDL73798 | conjugative transposon tetracycline resistance protein | - |
| Locus_33 | 34676..35611 | - | 936 | CDL73799 | putative conjugative transposon exported protein | orf13 |
| Locus_34 | 35608..36609 | - | 1002 | CDL73800 | putative cell wall hydrolase | orf14 |
| Locus_35 | 36606..38717 | - | 2112 | CDL73801 | conjugative transposon membrane protein | orf15 |
| Locus_36 | 38786..41110 | - | 2325 | CDL73802 | conjugative transposon ATP/GTP-binding protein | virb4 |
| Locus_37 | 41217..41447 | - | 231 | CDL73803 | putative conjugative transposon membrane protein | orf17a |
| Locus_38 | 41698..42195 | - | 498 | CDL73804 | conjugative transposon protein | - |
| Locus_39 | 42312..42479 | - | 168 | CDL73805 | conjugative transposon protein | orf19 |
| Locus_40 | 42576..43781 | - | 1206 | CDL73806 | putative conjugative transposon replication initiation factor | - |
| Locus_41 | 43959..45341 | - | 1383 | CDL73807 | conjugative transposon FtsK/SpoIIIE-family protein | virb4 |
| Locus_42 | 45370..45756 | - | 387 | CDL73808 | conjugative transposon protein | orf23 |
| Locus_43 | 45772..46059 | - | 288 | CDL73809 | conjugative transposon protein | orf23 |
| Locus_44 | 46392..47105 | + | 714 | CDL73810 | hypothetical protein | - |
| Locus_45 | 47068..47964 | - | 897 | CDL73811 | HTH-domain DNA binding protein | - |
| Locus_46 | 49127..49768 | + | 642 | CDL73812 | Conserved hypothetical protein | prgL |
| Locus_47 | 49778..50863 | + | 1086 | CDL73813 | conserved hypothetical protein | traP |
| Locus_48 | 50914..51147 | + | 234 | CDL73814 | conserved hypothetical protein | gbs1347 |
| Locus_49 | 51144..51527 | + | 384 | CDL73815 | conserved hypothetical protein | gbs1346 |
| Locus_50 | 51640..51840 | + | 201 | CDL73816 | conserved hypothetical protein | - |
| Locus_51 | 51922..52554 | + | 633 | CDL73817 | Conserved hypothetical protein | - |
| Locus_52 | 52545..52772 | + | 228 | CDL73818 | Conserved hypothetical protein | - |
| Locus_53 | 52893..53072 | - | 180 | CDL73819 | Conserved hypothetical protein | - |
| Locus_54 | 53142..53513 | + | 372 | CDL73820 | Conserved hypothetical protein | - |
| Locus_55 | 53890..54786 | - | 897 | CDL73821 | Integrase | - |
| Locus_56 | 54880..55452 | + | 573 | CDL73822 | Conserved hypothetical protein | - |
Host bacterium
| ID | 447 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICESpnDCC1738 | GenBank | HG799492 |
| Element size | 67809 bp | Coordinate of oriT [Strand] | 43792..43924 [-] |
| Host bacterium | Streptococcus pneumoniae integrative and | Coordinate of element | 1..67809 |
Cargo genes
| Drug resistance gene | tet(M) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21 |