Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200502 |
| Name | oriT_ICESpnDCC1524 |
| Organism | Streptococcus pneumoniae integrative and |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | HG799493 (19175..19307 [+], 133 nt) |
| oriT length | 133 nt |
| IRs (inverted repeats) | 65..70, 83..88 (AAATCC..GGATTT) 6..12, 23..29 (ACCCCCC..GGGGGGT) |
| Location of nic site | 75..76 |
| Conserved sequence flanking the nic site |
TTTGGTTACA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 133 nt
ACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file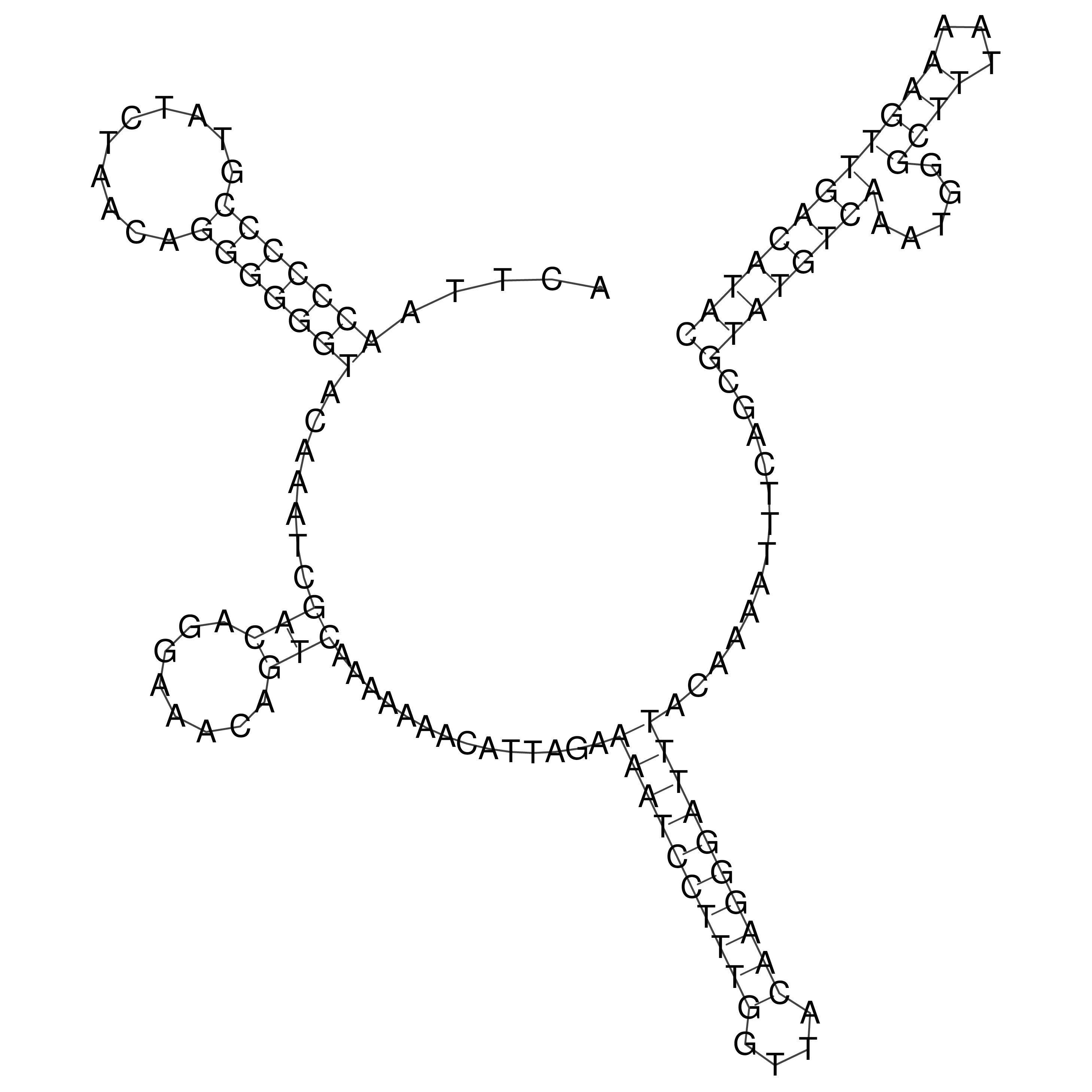
Relaxase
| ID | 14547 | GenBank | CDL73854 |
| Name | Rep_trans_-_ICESpnDCC1524 |
UniProt ID | _ |
| Length | 420 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 420 a.a. Molecular weight: 49663.70 Da Isoelectric Point: 7.3929
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKKGYLSTIPVDRYPKKRYNGR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 7923 | GenBank | CDL73868 |
| Name | CDL73868_ICESpnDCC1524 |
UniProt ID | _ |
| Length | 361 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 361 a.a. Molecular weight: 41696.88 Da Isoelectric Point: 9.6071
MATDRVPAGKRDCISLREKIAELQKDIHDGIDVVGKKMTLCQLYAKQNAQRPKVRKNTETGRKYLMDILK
KDKLGVRSIDSIKPSDAKEWAIRMSENGYAYQTINNYKRSLKASFYIAIQDDCVRKNPFDFQLKAVLDDD
TVPKTVLTEEQEEKLLAFAKADKTYSKNYDEILILLKTGLRISEFGGLTLPDLDFENRLVNIDHQLLRDT
EIGYYIETPKTKSGERQVPMVEEAYQAFKRVLANRKNDKRVEIDGYSDFLFLNRKNYPKVASDYNGMMKG
LVKKYNKYNEDKLPHITPHSLRHTFCTNYANAGMNPKALQYIMGHANIAMTLNYYAHATFDSAMAEMKRL
NKEKQQERLVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 17164 | GenBank | CDL73845 |
| Name | t4cp2_-_ICESpnDCC1524 |
UniProt ID | _ |
| Length | 626 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 626 a.a. Molecular weight: 72505.90 Da Isoelectric Point: 9.6194
MMYSGKKFLLFSLLGILLGYLFHRLTLLYDSYTGNTLDKWTHLLMEGQDEVLQSPWNVSFTGKSSAFFLL
GFVMMLLVYLYLETGKKQYREGVEYGSARFGTLKEKKLFYGKEFSHDTILAQDVRLTLLDKKPPQYDRNK
NIAVIGGSGSGKTFRFVKPNLIQMNSSNIVVDPKDHLAEKTGKLFLERGYQVKVLDLVNMKNSDGFNPFR
YIETENDLNRMLTVYFNNTKGSGSRSDPFWDEASMTLVRALASYLVDFYNPPKTREQLIEESRLSQKEYQ
NLLKRQKKEVEERKKRGRYPSFAEISKLIKHLSKGENQEKSVLEILFENYAKKYGTENFTMRNWADFQNY
KDKTLDSVIAVTTAKFALFNIQSVMDLTKRDTLDMKTWGKEKSMVYLVIPDNDSTFRFLSALFFSTVFQT
LTRQADIDFKGQLPLHVRVYLDEFANIGEIPDFAEQTSTVRSRNMSLVPILQNIAQLQGLYKEKEAWKTI
LGNCDSLVYLGGNDEDTFKFMSGLLGKQTIDVRNTSRSFGQTGSGSLSHQKIARDLMTPDEVGNMKRHEC
LVRIANMPVFKSKKYNSTKHPNWKYLANQENDERWWNYQINPLNQSQENHLEGLRIRDLTFESSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 17165 | GenBank | CDL73853 |
| Name | tcpA_-_ICESpnDCC1524 |
UniProt ID | _ |
| Length | 461 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 461 a.a. Molecular weight: 53370.27 Da Isoelectric Point: 9.0687
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKN
GLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVEAKDGKLRL
MKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLL
SCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLG
RQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSV
ISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 2391..42660
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| Locus_0 | 1..174 | + | 174 | CDL73831 | conserved hypothetical protein | - |
| Locus_1 | 171..950 | + | 780 | CDL73832 | putative replication initiator protein | - |
| Locus_2 | 1049..2407 | + | 1359 | CDL73833 | methyl transferase | - |
| Locus_3 | 1080..1622 | + | 543 | CDL73834 | methyl transferase | - |
| Locus_4 | 2391..2840 | + | 450 | CDL73835 | conserved hypothetical protein | gbs1369 |
| Locus_5 | 2833..3213 | + | 381 | CDL73836 | conserved hypothetical protein | - |
| Locus_6 | 3226..3459 | + | 234 | Protein_6 | putative uncharacterized protein | - |
| Locus_7 | 3462..4049 | + | 588 | CDL73838 | Putative membrane protein | - |
| Locus_8 | 4341..4976 | + | 636 | CDL73839 | hypothetical protein | - |
| Locus_9 | 5031..5273 | + | 243 | CDL73840 | caax amino protease family | - |
| Locus_10 | 5401..5991 | + | 591 | CDL73841 | conserved hypothetical protein | - |
| Locus_11 | 5991..6827 | + | 837 | CDL73842 | putative abortive infection protein AbiGII | - |
| Locus_12 | 7109..7408 | + | 300 | CDL73843 | conserved hypothetical protein | - |
| Locus_13 | 7422..7886 | + | 465 | CDL73844 | conserved hypothetical protein | gbs1365 |
| Locus_14 | 7883..9763 | + | 1881 | CDL73845 | putative conjugal transfer protein TraG | virb4 |
| Locus_15 | 9784..10026 | + | 243 | CDL73846 | conserved hypothetical protein | prgF |
| Locus_16 | 10043..10897 | + | 855 | CDL73847 | membrane protein, putative | prgHb |
| Locus_17 | 10951..11310 | + | 360 | CDL73848 | conserved hypothetical protein | prgIc |
| Locus_18 | 11303..13618 | + | 2316 | CDL73849 | putative conjugal transfer protein | virb4 |
| Locus_19 | 13630..16443 | + | 2814 | CDL73850 | putative conjugal transfer protein | prgK |
| Locus_20 | 17037..17324 | + | 288 | CDL73851 | conjugative transposon protein | orf23 |
| Locus_21 | 17340..17726 | + | 387 | CDL73852 | conjugative transposon protein | orf23 |
| Locus_22 | 17755..19140 | + | 1386 | CDL73853 | conjugative transposon FtsK/SpoIIIE-family protein | virb4 |
| Locus_23 | 19318..20580 | + | 1263 | CDL73854 | putative conjugative transposon replication initiation factor | - |
| Locus_24 | 20994..21731 | + | 738 | CDL73855 | rRNA adenine N-6-methyltransferase | - |
| Locus_25 | 21988..23235 | - | 1248 | CDL73856 | transposase | - |
| Locus_26 | 23415..23636 | + | 222 | CDL73857 | conjugative transposon protein | orf19 |
| Locus_27 | 23753..24250 | + | 498 | CDL73858 | conjugative transposon protein | - |
| Locus_28 | 24501..24731 | + | 231 | CDL73859 | putative conjugative transposon membrane protein | orf17a |
| Locus_29 | 24838..27162 | + | 2325 | CDL73860 | conjugative transposon ATP/GTP-binding protein | virb4 |
| Locus_30 | 27231..29342 | + | 2112 | CDL73861 | conjugative transposon membrane protein | orf15 |
| Locus_31 | 29339..30340 | + | 1002 | CDL73862 | putative cell wall hydrolase | orf14 |
| Locus_32 | 30337..31269 | + | 933 | CDL73863 | putative conjugative transposon exported protein | orf13 |
| Locus_33 | 31631..33565 | + | 1935 | CDL73864 | conjugative transposon tetracycline resistance protein | - |
| Locus_34 | 33911..34264 | - | 354 | CDL73865 | putative conjugative transposon regulatory protein | - |
| Locus_35 | 34769..35191 | + | 423 | CDL73866 | putative conjugative transposon regulatory protein | - |
| Locus_36 | 35879..36082 | + | 204 | CDL73867 | excisionase | - |
| Locus_37 | 36296..37381 | + | 1086 | CDL73868 | integrase | - |
| Locus_38 | 37558..38238 | + | 681 | CDL73869 | hypothetical protein | - |
| Locus_39 | 38201..39097 | - | 897 | CDL73870 | HTH-domain DNA binding protein | - |
| Locus_40 | 39376..39564 | - | 189 | CDL73871 | Hypothetical protein | - |
| Locus_41 | 40260..40901 | + | 642 | CDL73872 | Conserved hypothetical protein | prgL |
| Locus_42 | 40911..41996 | + | 1086 | CDL73873 | conserved hypothetical protein | traP |
| Locus_43 | 42047..42280 | + | 234 | CDL73874 | conserved hypothetical protein | gbs1347 |
| Locus_44 | 42277..42660 | + | 384 | CDL73875 | conserved hypothetical protein | gbs1346 |
| Locus_45 | 42773..42973 | + | 201 | CDL73876 | conserved hypothetical protein | - |
| Locus_46 | 43055..43687 | + | 633 | CDL73877 | Conserved hypothetical protein | - |
| Locus_47 | 43678..43905 | + | 228 | CDL73878 | Conserved hypothetical protein | - |
| Locus_48 | 44026..44205 | - | 180 | CDL73879 | Conserved hypothetical protein | - |
| Locus_49 | 44275..44646 | + | 372 | CDL73880 | Conserved hypothetical protein | - |
| Locus_50 | 45023..45919 | - | 897 | CDL73881 | Integrase | - |
| Locus_51 | 46013..46585 | + | 573 | CDL73882 | Conserved hypothetical protein | - |
Host bacterium
| ID | 446 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICESpnDCC1524 | GenBank | HG799493 |
| Element size | 58942 bp | Coordinate of oriT [Strand] | 19175..19307 [+] |
| Host bacterium | Streptococcus pneumoniae integrative and | Coordinate of element | 1..58942 |
Cargo genes
| Drug resistance gene | erm(B), tet(M) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21 |