Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200501 |
| Name | oriT_ICESpn6BST273 |
| Organism | Streptococcus pneumoniae integrative and |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | HG799495 (44930..45062 [+], 133 nt) |
| oriT length | 133 nt |
| IRs (inverted repeats) | 65..70, 83..88 (AAATCC..GGATTT) 6..12, 23..29 (ACCCCCC..GGGGGGT) |
| Location of nic site | 75..76 |
| Conserved sequence flanking the nic site |
TTTGGTTACA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 133 nt
ACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file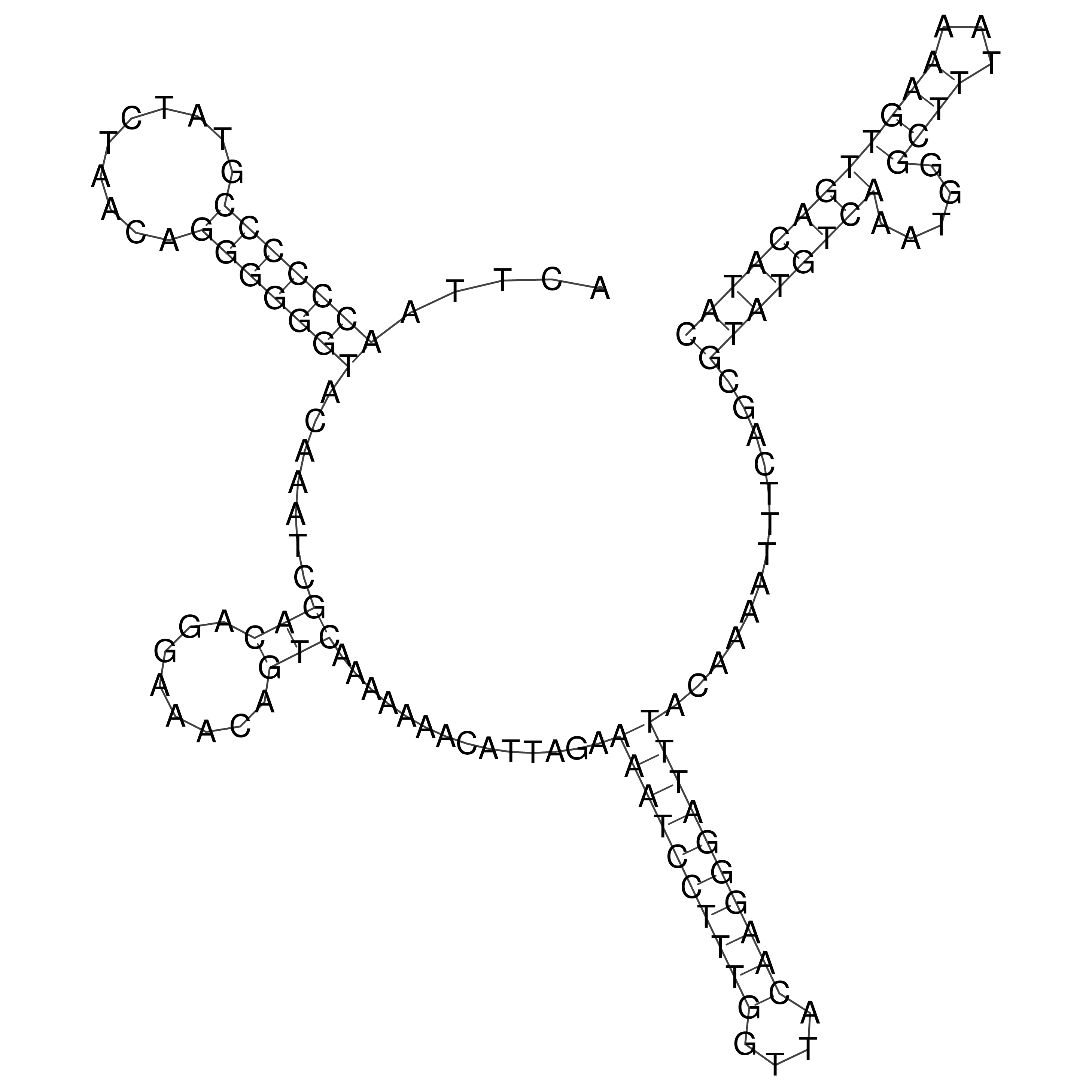
Relaxase
| ID | 14545 | GenBank | CDL73932 |
| Name | Relaxase_-_ICESpn6BST273 |
UniProt ID | _ |
| Length | 607 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 607 a.a. Molecular weight: 73076.29 Da Isoelectric Point: 6.6667
MVITKHFAIHGKSYRRKLIKYILNPDKTKNLTLVSDYDMRNFLDFPSYEEMVQMYHENFINNDTLYNFRH
DRLEEKQRQIHAHHIIQSFSPEDNLTPEQINQIGYETMKELTSGKFRFIVATHVDKDHLHNHIIINSVDS
NSDKKLKWDYKVERNLRMISDRFSKIAGAKIIENRYSHQQYEVYRKTNHKYELKQRLYFLMEHSRDFEDF
KKNAPLLHVEMDFSHKHATFFMTDSSMKQVVRGNKLNRKQPYTEEFFKNYFAKREMEELMEFLLSKAENI
DDLIETAKLFDLTIIPKQKHVYFQLAGVEVKETELDQKNLYGVEFFQDYFESRKDWQNPEIEDLVQFYQE
EKFSKEKELPRDEKFWEFYQEFKSNRDAVHEFEVELSLNQIEKVVDDGIYIKVKFGIRQEGLIFVPNMQL
DIEEEKVKVFIRETSSYYVYHKDDPEKNRYMKGRTLIRQFSSENQTIPFRRKTTVDMIKEKIEEVYTLLE
LDMDKHSYITIKDDLIHELAASELRINALQERVTTLNQVAEYLLASVENKQKMKLNLSKLNITDKIGIDV
VEKELNELGIQIELESNRYENIVFKVNKFINRLSKKISKEEGICFQK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 14546 | GenBank | CDL73939 |
| Name | Rep_trans_-_ICESpn6BST273 |
UniProt ID | _ |
| Length | 401 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 401 a.a. Molecular weight: 47398.10 Da Isoelectric Point: 6.5091
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 7922 | GenBank | CDL73954 |
| Name | CDL73954_ICESpn6BST273 |
UniProt ID | _ |
| Length | 361 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 361 a.a. Molecular weight: 41696.88 Da Isoelectric Point: 9.6071
MATDRVPAGKRDCISLREKIAELQKDIHDGIDVVGKKMTLCQLYAKQNAQRPKVRKNTETGRKYLMDILK
KDKLGVRSIDSIKPSDAKEWAIRMSENGYAYQTINNYKRSLKASFYIAIQDDCVRKNPFDFQLKAVLDDD
TVPKTVLTEEQEEKLLAFAKADKTYSKNYDEILILLKTGLRISEFGGLTLPDLDFENRLVNIDHQLLRDT
EIGYYIETPKTKSGERQVPMVEEAYQAFKRVLANRKNDKRVEIDGYSDFLFLNRKNYPKVASDYNGMMKG
LVKKYNKYNEDKLPHITPHSLRHTFCTNYANAGMNPKALQYIMGHANIAMTLNYYAHATFDSAMAEMKRL
NKEKQQERLVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 17162 | GenBank | CDL73902 |
| Name | t4cp2_-_ICESpn6BST273 |
UniProt ID | _ |
| Length | 625 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 625 a.a. Molecular weight: 72254.60 Da Isoelectric Point: 9.6195
MYSGKKFLLFSLLGILLGYLFHRLTLLYDSYTGNILDKWTHLLMEGQDEVLQSPWNVSFTGKSSAFFLLG
FVMMLLVYLYLETGKKQYREGVEYGSARFGTLKEKKLFYGKEFSHDTILAQDVRLTLLDKKPPQYDRNKN
IAVIGGSGSGKTFRFVKPNLIQMNSSNIVVDPKDHLAEKTGKLFLEHGYQVKVLDLVNMNNSDGFNPFRY
IETENDLNRMLTVYFNNTKGSGSRSDPFWDEASMTLVRALASYLVDFYNPPKTREQLIKESRLSQKEYQN
LLKGQKKEVEERKKRGRYPSFAEISKLIKHLSKGENQDKSVLEILFENYAKKYGTENFTMRNWADFQNYK
DKTLDSVVAVTTAKFALFNIQSVMDLTKRDTLDMKTWGKEKSMVYLVIPDNDSTFRFLSALFFSTVFQTL
TRQADIDFKGQLPLHVRVYLDEFANIGEIPDFAEQTSTVRSRNMSLVPILQNIAQLQGLYKEKEAWKTIL
GNCDSLVYLGGNDEDTFKFMSGLLGKQTIDVRNTSRSFGQTGSGSLSHQKIARDLMTPDEVGNMKRHECL
VRIANMPVFKSKKYSSTKHPNWKYLANQETDERWWNYQINPLNQRQENHLEGLRIRDLTFESSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 17163 | GenBank | CDL73938 |
| Name | tcpA_-_ICESpn6BST273 |
UniProt ID | _ |
| Length | 407 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 407 a.a. Molecular weight: 47303.95 Da Isoelectric Point: 9.2739
MKQRGKRIRPSGKDLVFHFTIAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRT
KEKITYFPKMYYRLKNGLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIAS
RISIDEVEAKDGKLRLMKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLAD
LGSVMANVYYRKEDLLSCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTK
ENTAVMNKLKQIVMLGRQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFF
LKRIKGRGYVDVGTSVISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 2431..26625
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| Locus_0 | 1..213 | + | 213 | CDL73891 | putative uncharacterized protein | - |
| Locus_1 | 210..989 | + | 780 | CDL73892 | putative replication initiator protein | - |
| Locus_2 | 1089..2447 | + | 1359 | CDL73893 | putative DNA methylase | - |
| Locus_3 | 2431..2880 | + | 450 | CDL73894 | putative uncharacterized protein | gbs1369 |
| Locus_4 | 2873..3253 | + | 381 | CDL73895 | putative uncharacterized protein | - |
| Locus_5 | 3266..3499 | + | 234 | CDL73896 | putative uncharacterized protein | - |
| Locus_6 | 3502..4089 | + | 588 | CDL73897 | putative CAAX amino terminal protease | - |
| Locus_7 | 4220..4810 | + | 591 | CDL73898 | conserved hypothetical protein | - |
| Locus_8 | 4810..5646 | + | 837 | CDL73899 | conserved hypothetical protein | - |
| Locus_9 | 5929..6228 | + | 300 | CDL73900 | putative uncharacterized protein | - |
| Locus_10 | 6242..6706 | + | 465 | CDL73901 | putative uncharacterized protein | gbs1365 |
| Locus_11 | 6706..8583 | + | 1878 | CDL73902 | putative conjugal transfer protein TraG | virb4 |
| Locus_12 | 8604..8846 | + | 243 | CDL73903 | putative uncharacterized protein | prgF |
| Locus_13 | 8863..9717 | + | 855 | CDL73904 | putative uncharacterized protein | prgHb |
| Locus_14 | 9771..10130 | + | 360 | CDL73905 | putative uncharacterized protein | prgIc |
| Locus_15 | 10123..12438 | + | 2316 | CDL73906 | putative conjugal transfer protein | virb4 |
| Locus_16 | 12450..15263 | + | 2814 | CDL73907 | putative conjugal transfer protein | prgK |
| Locus_17 | 15298..15906 | - | 609 | CDL73908 | Conserved hypothetical protein | - |
| Locus_18 | 15908..16072 | - | 165 | CDL73909 | Conserved hypothetical protein | - |
| Locus_19 | 16128..16421 | - | 294 | CDL73910 | Conserved hypothetical protein | - |
| Locus_20 | 16533..16943 | + | 411 | Protein_20 | putative uncharacterized protein | - |
| Locus_21 | 16976..23209 | + | 6234 | CDL73912 | putative conjugative transposon DNA recombination protein | - |
| Locus_22 | 23284..23568 | + | 285 | CDL73913 | putative uncharacterized protein | - |
| Locus_23 | 24225..24866 | + | 642 | CDL73914 | putative uncharacterized protein | prgL |
| Locus_24 | 24876..25961 | + | 1086 | CDL73915 | putative uncharacterized protein | traP |
| Locus_25 | 26012..26245 | + | 234 | CDL73916 | putative uncharacterized protein | gbs1347 |
| Locus_26 | 26242..26625 | + | 384 | CDL73917 | putative uncharacterized protein | gbs1346 |
| Locus_27 | 26738..26935 | + | 198 | CDL73918 | putative uncharacterized protein | - |
| Locus_28 | 27093..27479 | + | 387 | CDL73919 | putative epsilon antitoxin | - |
| Locus_29 | 27479..28231 | + | 753 | CDL73920 | zeta toxin | - |
| Locus_30 | 28421..28558 | + | 138 | CDL73921 | Conserved hypothetical protein | - |
| Locus_31 | 28616..28828 | + | 213 | CDL73922 | Conserved hypothetical protein | - |
| Locus_32 | 28849..29055 | + | 207 | CDL73923 | Conserved hypothetical protein | - |
| Locus_33 | 29252..30148 | - | 897 | CDL73924 | putative integrase | - |
| Locus_34 | 30242..30814 | + | 573 | CDL73925 | Conserved hypothetical protein | - |
Region 2: 42954..54175
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| Locus_39 | 38208..38564 | + | 357 | CDL73930 | Tn5252 orf10 protein | - |
| Locus_40 | 38569..38934 | + | 366 | CDL73931 | Tn5252 orf9 protein | - |
| Locus_41 | 38921..40744 | + | 1824 | CDL73932 | Tn5252 relaxase | - |
| Locus_42 | 40948..41196 | + | 249 | CDL73933 | Conserved hypothetical protein | - |
| Locus_43 | 41301..42164 | - | 864 | CDL73934 | putative transcriptional regulator | - |
| Locus_44 | 42355..42525 | + | 171 | CDL73935 | putative secreted protein | - |
| Locus_45 | 42954..43241 | + | 288 | CDL73936 | conjugative transposon protein | orf23 |
| Locus_46 | 43257..43643 | + | 387 | CDL73937 | conjugative transposon protein | orf23 |
| Locus_47 | 43672..44895 | + | 1224 | CDL73938 | conjugative transposon FtsK/SpoIIIE-family protein | virb4 |
| Locus_48 | 45073..46278 | + | 1206 | CDL73939 | putative conjugative transposon replication initiation factor | - |
| Locus_49 | 46321..46542 | + | 222 | CDL73940 | conjugative transposon protein | orf19 |
| Locus_50 | 46659..47156 | + | 498 | CDL73941 | conjugative transposon protein | - |
| Locus_51 | 47407..47637 | + | 231 | CDL73942 | putative conjugative transposon membrane protein | orf17a |
| Locus_52 | 47744..50068 | + | 2325 | CDL73943 | conjugative transposon ATP/GTP-binding protein | virb4 |
| Locus_53 | 50137..52248 | + | 2112 | CDL73944 | conjugative transposon membrane protein | orf15 |
| Locus_54 | 52245..53246 | + | 1002 | CDL73945 | putative cell wall hydrolase | orf14 |
| Locus_55 | 53243..54175 | + | 933 | CDL73946 | putative conjugative transposon exported protein | orf13 |
| Locus_56 | 54537..56471 | + | 1935 | CDL73947 | conjugative transposon tetracycline resistance protein | - |
| Locus_58 | 57578..58315 | + | 738 | CDL73949 | rRNA methylase | - |
Host bacterium
| ID | 445 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICESpn6BST273 | GenBank | HG799495 |
| Element size | 76642 bp | Coordinate of oriT [Strand] | 44930..45062 [+] |
| Host bacterium | Streptococcus pneumoniae integrative and | Coordinate of element | 1..76642 |
Cargo genes
| Drug resistance gene | tet(M), erm(B) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21, AcrIB |