Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200499 |
| Name | oriT2_ICESluvan |
| Organism | Enterococcus faecium transfer chromosomal element strain MM5-F9a |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | HE963029 ( 36534..36563 [-], 30 nt) |
| oriT length | 30 nt |
| IRs (inverted repeats) | 1..8, 13..20 (GGGAGTGT..ACACTCCC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 30 nt
GGGAGTGTAGCCACACTCCCTATGCTTGCT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file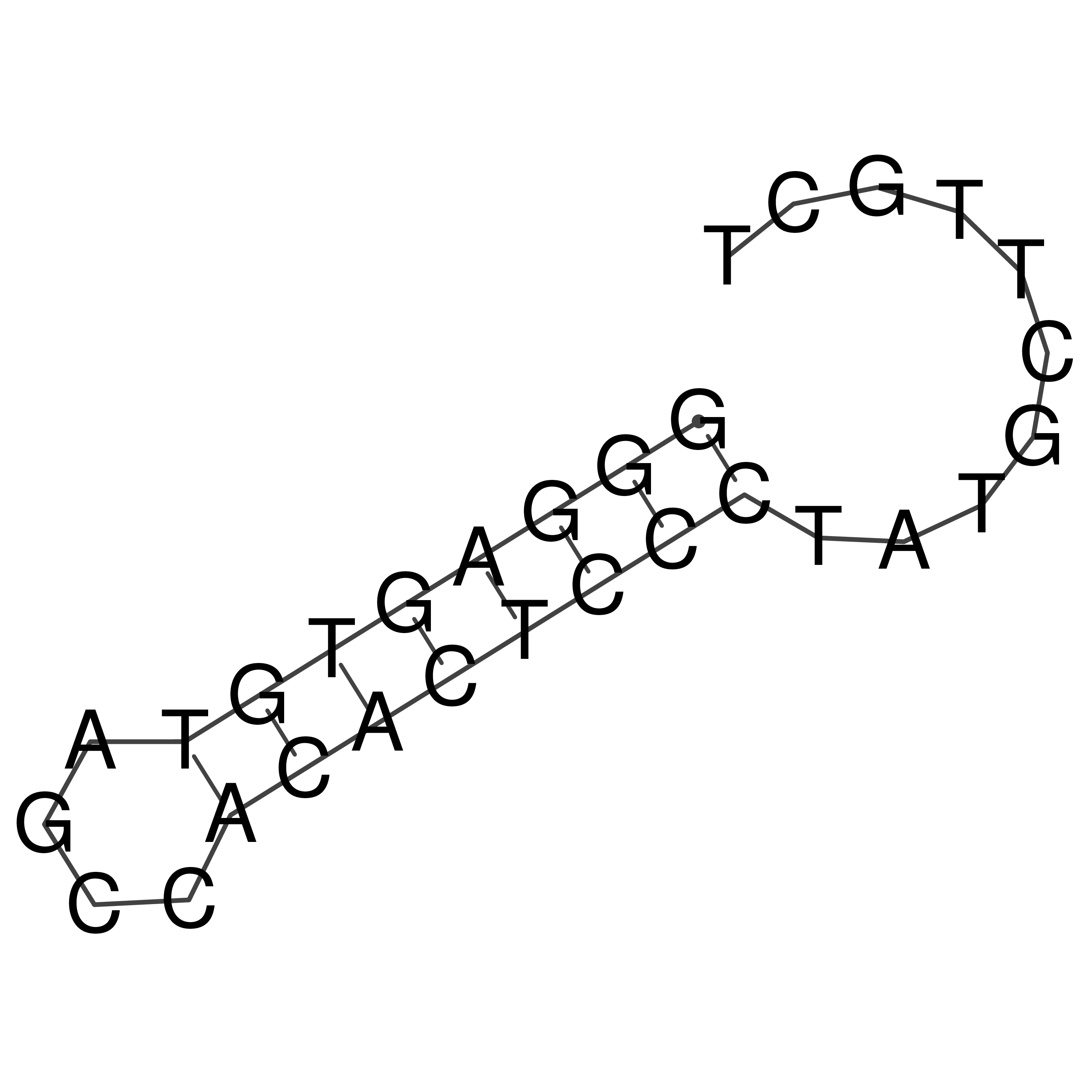
Relaxase
| ID | 14540 | GenBank | CCJ27760 |
| Name | MobA_MobL_-_ICESluvan |
UniProt ID | _ |
| Length | 502 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 502 a.a. Molecular weight: 57294.96 Da Isoelectric Point: 9.4448
MIPIAIYHCNISIVSRGKGKSAVAAAAYRSGEKLTNEWDGMAHDYTRKGGVVHTEIMLPPHAPPSFSDRS
TLWNSVELYEKAGNAQLAREIDAALPIELSREEQIRLVREYCSSQFVSRGMCVDYAIHDTGKGNPHCHIM
LTMRPLDERGAWAAKSRKEYDLDENGERIRLPSGRYKTHKVDLTGWNDKGNALLWRKAWADLTNDFLERN
GSPQRIDHRSNAERGIDEIPTVHMGVAACQMEKKGIATEKGELNRNIQKANRLIREIRAQVGKLKEWIAD
LFKARETAPEQPPQSPGLANLLMKYLGVQREKSRKYSQSWQRQHAADELKTVAAAVNYLSEHGISTLDEL
DAALSSVSEQADAIRAGMKTAEQRMKELQKLIEYGRNYQTYKPVQDEYRQIRWKGKQEKFAEARRTELTL
WNAANRYLHAHLPEGVKTLPISAWEKEFAALKAQREMEYAKLKGARSDLSELQNIRRCVDIALRADQPEQ
TQTRTKRHDIDR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 14541 | GenBank | CCJ27767 |
| Name | Relaxase_-_ICESluvan |
UniProt ID | _ |
| Length | 442 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 442 a.a. Molecular weight: 50173.57 Da Isoelectric Point: 9.6754
MAVTKIHPIKSTLKKALDYIENPDKTDGKLLVASFACSYETADIEFEMLLAQAYQKGNNLGHHLIQSFAP
GEATPEQAHEIGKQLAEEVLQGKYPYVLTTHIDKGHVHNHIIFCAVDMVNQRKYISNKQSYAYIRRTSDR
LCKENGLSVVKPGQSKGKSYAEWDAQRKGTSWKAKLKAAIDAAIPQAKDFDDFLRLMQAQGYEIKPGKFI
SFRAPGQERFTRCKTLGEAYTEEAIKERIKGRVITRAPKERKGISLRIDLENNIKAQQSAGYERWAKLHN
LKQAAKTLNFLTEHGIDTYPDLESKVAEITAASDEAAATLKAMEHRLADMAVLIKNISTYKQLWPVAMEY
RNAADKAKFRREHESTLILYEAAAKALKGQGVKKLPDLYALKAEYKRLAEEKERLYEKYGEAKKQMQEYG
IIKQNVDGILRMTPGKEWTQEL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 7921 | GenBank | CCJ27768 |
| Name | CCJ27768_ICESluvan |
UniProt ID | _ |
| Length | 109 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 109 a.a. Molecular weight: 12486.63 Da Isoelectric Point: 10.5610
MANRKRKIVLRVPVTPEERAMIEQKMALLHTKNFSAYARKMLIDGYIVNMDTSDIRAQTAEIQKIGVNVN
QIAKRLNGMGPVYAQDIEDIKGALAQIWQLQRYILSSQR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 17159 | GenBank | CCJ27745 |
| Name | t4cp2_-_ICESluvan |
UniProt ID | _ |
| Length | 564 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 564 a.a. Molecular weight: 63615.31 Da Isoelectric Point: 7.2685
MKPDVKKLLILNLPYLIFVYLFAKCGEAYRLAAGADLSGKLLHFADGFAAAFANPLPSLHLFDLCIGIAG
AVIVRLVVYCKSKNVKKYRKGMEYGSARWGTAKDIAPYIDPKFENNILLTQTERLTMTGRPKDPKTARNK
NVLVIGGSGSGKTRFFVKPNLMQCFPTTDYPTSFVVTDPKGTLVLETGRMFQQAGYRIKILNTINFSKSM
KYNPFVYIHSEKDVLKLVNTLISNTKGEGEKSAEDFWVKSERLFYSALIGYIWYEAPAEEMNFSTLLEMI
NASEAREDDPEFQSPVDLMFERLEQKDPDHFAVRQYKKFLLSAGKTRSSILISCGARLAPFDIREVRELM
EDDEMELDTIGDEKTVLYLIMSDTDTTFNFILAMLQSQLINLLCDRADDVYGGRLPVHVRLILDEFANIG
QIPNFDKLIATIRSREISASIILQSQSQLKAIYKDAAEIISDNCDSVLFLSGRGKNAKEISDALGKETID
SFNTSENRGSQTSHGLNYQKLGKELMSQDEIAVMDGGKCILQLRGVRPVFIGEIRYYQASPVQIPCRRRQ
EEYL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 2729..35527
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| Locus_0 | 1..42 | + | 42 | Protein_0 | 23S rRNA (uracil-5-)-methyltransferase RumA (fragment) | - |
| Locus_1 | 9..176 | + | 168 | CCJ27726 | putative uncharacterized protein | - |
| Locus_2 | 253..426 | + | 174 | CCJ27727 | putative uncharacterized protein | - |
| Locus_3 | 423..1241 | + | 819 | CCJ27728 | replication initiator protein | - |
| Locus_4 | 1390..2745 | + | 1356 | CCJ27729 | cytosine-specific methyltransferase | - |
| Locus_5 | 2729..3157 | + | 429 | CCJ27730 | putative uncharacterized protein | gbs1369 |
| Locus_6 | 3168..3551 | + | 384 | CCJ27731 | putative uncharacterized protein | - |
| Locus_7 | 3560..3793 | + | 234 | CCJ27732 | putative uncharacterized protein | - |
| Locus_8 | 3796..4380 | + | 585 | CCJ27733 | CAAX amino terminal protease self-immunity protein | - |
| Locus_9 | 4421..4945 | + | 525 | CCJ27734 | putative uncharacterized protein | gbs1365 |
| Locus_10 | 4945..6761 | + | 1817 | Protein_10 | conjugal transfer protein TraG | - |
| Locus_11 | 6767..7021 | + | 255 | CCJ27736 | putative membrane protein | prgF |
| Locus_12 | 7045..7899 | + | 855 | CCJ27737 | putative plasmid conjugal transfer protein,TrbL/VirB6 family | prgHb |
| Locus_13 | 7859..8314 | + | 456 | CCJ27738 | putative uncharacterized protein | prgIc |
| Locus_14 | 8292..10622 | + | 2331 | CCJ27739 | putative conjugal transfer protein | virb4 |
| Locus_15 | 10624..13425 | + | 2802 | CCJ27740 | N-acetylmuramoyl L-alanine amidase | prgK |
| Locus_17 | 13838..15037 | + | 1200 | CCJ27742 | putative uncharacterized protein | - |
| Locus_18 | 15059..15271 | + | 213 | CCJ27743 | putative uncharacterized protein | - |
| Locus_19 | 15346..15822 | + | 477 | CCJ27744 | putative uncharacterized protein | cd411 |
| Locus_20 | 15819..17513 | + | 1695 | CCJ27745 | putative conjugal transfer protein, TraG/TraD family | - |
| Locus_21 | 17672..17920 | + | 249 | CCJ27746 | putative conjugal transfer protein | prgF |
| Locus_22 | 18003..18875 | + | 873 | CCJ27747 | membrane protein, putative conjugative transfer protein | prgHa |
| Locus_23 | 18906..19448 | + | 543 | CCJ27748 | adenine-specific methyltransferase | - |
| Locus_24 | 19462..19884 | + | 423 | CCJ27749 | putative uncharacterized protein | prgIa |
| Locus_25 | 19814..22213 | + | 2400 | CCJ27750 | putative conjugal transfer protein | virb4 |
| Locus_26 | 22245..24236 | + | 1992 | CCJ27751 | DNA-repair protein | cd419a |
| Locus_27 | 24259..24510 | + | 252 | CCJ27752 | putative uncharacterized protein | - |
| Locus_28 | 24500..25729 | + | 1230 | CCJ27753 | putative bacteriocin | - |
| Locus_29 | 25726..36520 | + | 10795 | CCJ27754 | DNA topoisomerase (pseudogene) | - |
| Locus_30 | 27204..29066 | + | 1863 | CCJ27755 | TnpX site-specific recombinase | - |
| Locus_31 | 29253..29549 | + | 297 | CCJ27756 | putative uncharacterized protein | - |
| Locus_32 | 29680..30733 | + | 1054 | CCJ27757 | transposase, IS30 family (pseudogene) | - |
| Locus_33 | 30982..31470 | + | 489 | CCJ27758 | putative VanZ family protein | - |
| Locus_34 | 31555..31881 | + | 327 | CCJ27759 | putative uncharacterized protein | - |
| Locus_35 | 32121..33629 | + | 1509 | CCJ27760 | putative mobilization protein, conjugal transfer protein | - |
| Locus_36 | 33626..35527 | + | 1902 | CCJ27761 | putative DNA primase | traP |
| Locus_37 | 35646..35837 | + | 192 | CCJ27762 | putative uncharacterized protein | - |
Region 2: 57625..78120
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| Locus_54 | 53022..53798 | + | 777 | CCJ27779 | putative uncharacterized protein | - |
| Locus_55 | 53805..54038 | + | 234 | CCJ27780 | putative uncharacterized protein | - |
| Locus_56 | 53960..54202 | - | 243 | CCJ27781 | putative uncharacterized protein | - |
| Locus_57 | 54462..54662 | + | 201 | CCJ27782 | excisionase | - |
| Locus_58 | 54665..55939 | + | 1275 | CCJ27783 | integrase | - |
| Locus_59 | 56862..57452 | - | 591 | CCJ27784 | putative abortive infection protein AbiGI | - |
| Locus_60 | 57625..62520 | + | 4896 | CCJ27785 | agglutinin receptor | prgB |
| Locus_61 | 62521..62712 | + | 192 | CCJ27786 | putative calcium-binding protein | - |
| Locus_62 | 62696..63247 | + | 552 | CCJ27787 | putative uncharacterized protein | gbs1354 |
| Locus_63 | 63296..70098 | + | 6803 | Protein_63 | SNF2-related helicase (pseudogene) | - |
| Locus_64 | 70169..70468 | + | 300 | CCJ27789 | putative uncharacterized protein | - |
| Locus_65 | 70482..70781 | + | 300 | CCJ27790 | putative uncharacterized protein | gbs1350 |
| Locus_66 | 71481..72686 | + | 1206 | CCJ27791 | putative uncharacterized phage related protein | - |
| Locus_67 | 72683..74011 | + | 1329 | CCJ27792 | putative uncharacterized phage related protein | - |
| Locus_68 | 74246..75503 | - | 1258 | Protein_68 | transposase, ISL3 family (pseudogene) | - |
| Locus_69 | 75699..76337 | + | 639 | CCJ27794 | putative uncharacterized protein | prgL |
| Locus_70 | 76377..77453 | + | 1077 | CCJ27795 | putative DNA primase | - |
| Locus_71 | 77507..77734 | + | 228 | CCJ27796 | putative uncharacterized protein | gbs1347 |
| Locus_72 | 77731..78120 | + | 390 | CCJ27797 | putative uncharacterized protein | gbs1346 |
| Locus_73 | 78170..78460 | + | 291 | CCJ27798 | putative uncharacterized protein | - |
| Locus_74 | 78500..79006 | + | 507 | CCJ27799 | putative antidote epsilon protein, zeta toxin regulator | - |
| Locus_75 | 79006..79776 | + | 771 | CCJ27800 | putative zeta-toxin/signal recognition particle | - |
| Locus_76 | 79808..82315 | + | 2508 | CCJ27801 | putative uncharacterized protein | - |
| Locus_77 | 82636..82995 | + | 360 | CCJ27802 | putative uncharacterized protein | - |
Host bacterium
| ID | 443 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICESluvan | GenBank | HE963029 |
| Element size | 94189 bp | Coordinate of oriT [Strand] | 44538..44565 [+]; 36534..36563 [-] |
| Host bacterium | Enterococcus faecium transfer chromosomal element strain MM5-F9a | Coordinate of element | 1..94189 |
Cargo genes
| Drug resistance gene | VanHBX |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21 |