Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200496 |
| Name | oriT_ICEBpSAFR-032 |
| Organism | Bacillus pumilus SAFR-032 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NC_009848 (6282..6305 [+], 24 nt) |
| oriT length | 24 nt |
| IRs (inverted repeats) | 1..7, 18..24 (ACCCCCC..GGGGGGT) 3..8, 18..23 (CCCCCC..GGGGGG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 24 nt
>oriT_ICEBpSAFR-032
ACCCCCCCACGCTAACAGGGGGGT
ACCCCCCCACGCTAACAGGGGGGT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file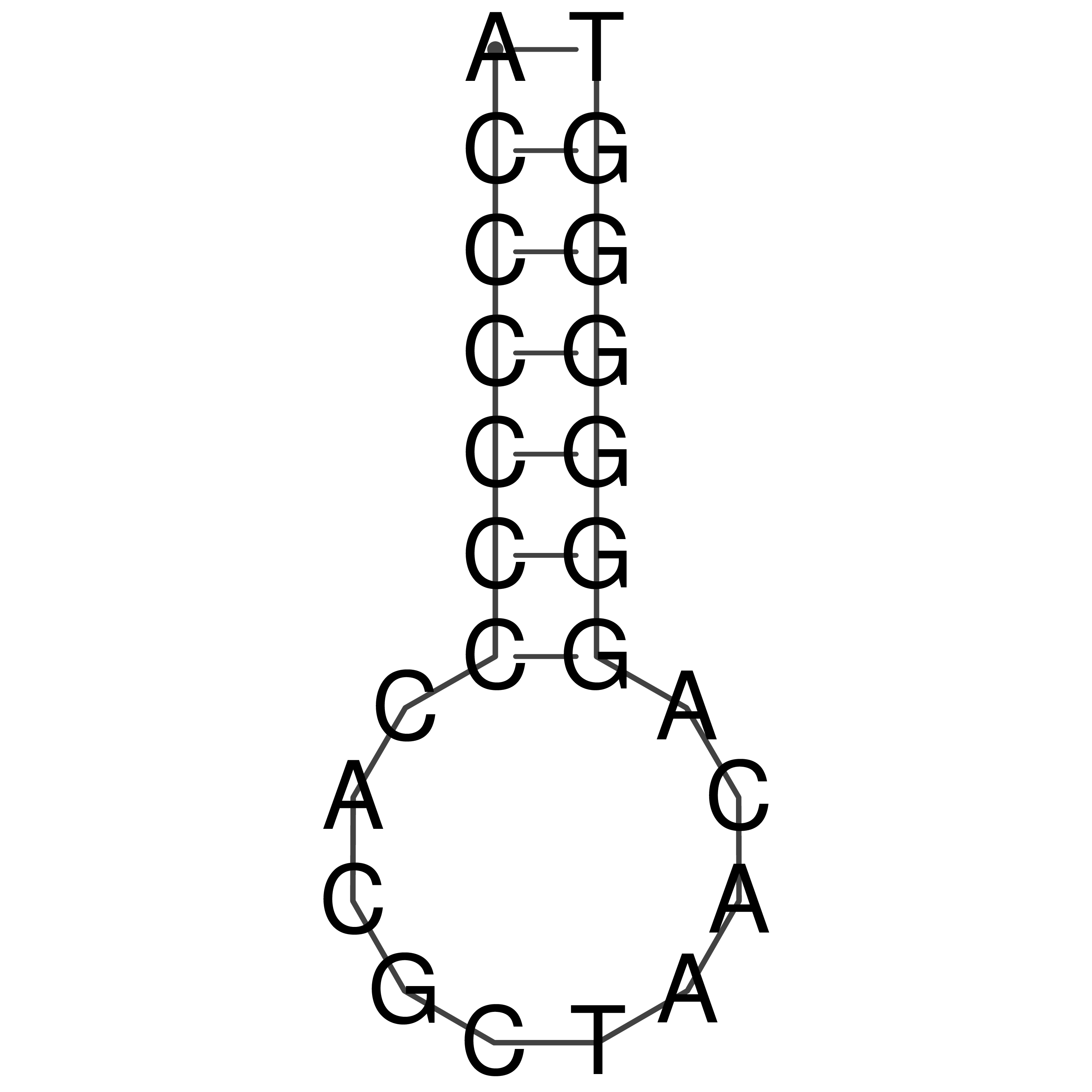
Relaxase
| ID | 14536 | GenBank | WP_012009099 |
| Name | Rep_trans_BPUM_RS02920_ICEBpSAFR-032 |
UniProt ID | A8FAI1 |
| Length | 366 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 366 a.a. Molecular weight: 42620.80 Da Isoelectric Point: 8.2711
>WP_012009099.1 replication initiation factor domain-containing protein [Bacillus pumilus]
MTGKVKPPHANRGVLRKKEEEKSQLTSMVDYVRISFKTHDIDHIIQNVLHINREFMTEEPRGFYGYIGTW
RMDMIKVFYSAPDDERGTLIELSGKGCRQFESFLECRGKTWFDFFQDCLNEKGNFTRLDIAIDDKKTYFS
IPNLLKRVQKGLCVSRYRRVDNNGSNQIEDGSSAGTTIYFGSKKSESYLCFYEKNYEQADKFNLSYDDLE
KWNRYEIRLKSERAHSAVCALIADRNLKKIAIGIISNCIRFVEEKGEGDKRKRKTSPFWLKFIGDVEKLQ
LTIEPKKDFYEKSRNWLQNSCAPTMKVILEVDRVLGNSRLLDVILNAEEGALGDSDLLNMIFNAKLQDKH
EKMLETFLTPIHEMVV
MTGKVKPPHANRGVLRKKEEEKSQLTSMVDYVRISFKTHDIDHIIQNVLHINREFMTEEPRGFYGYIGTW
RMDMIKVFYSAPDDERGTLIELSGKGCRQFESFLECRGKTWFDFFQDCLNEKGNFTRLDIAIDDKKTYFS
IPNLLKRVQKGLCVSRYRRVDNNGSNQIEDGSSAGTTIYFGSKKSESYLCFYEKNYEQADKFNLSYDDLE
KWNRYEIRLKSERAHSAVCALIADRNLKKIAIGIISNCIRFVEEKGEGDKRKRKTSPFWLKFIGDVEKLQ
LTIEPKKDFYEKSRNWLQNSCAPTMKVILEVDRVLGNSRLLDVILNAEEGALGDSDLLNMIFNAKLQDKH
EKMLETFLTPIHEMVV
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A8FAI1 |
T4CP
| ID | 17155 | GenBank | WP_275477915 |
| Name | tcpA_BPUM_RS02915_ICEBpSAFR-032 |
UniProt ID | _ |
| Length | 417 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 417 a.a. Molecular weight: 48548.64 Da Isoelectric Point: 9.3235
>WP_275477915.1 FtsK/SpoIIIE domain-containing protein [Bacillus pumilus]
MPVFLLLMILFWRSHLWDVYEKGIEDTFENFTFNIPLIVKSLLVSIIVPVALILVWYFIFHQSYKKIYHR
QKIARMIFSNKFYESKSVRSKITEKTVQKISYFPKFYYRVKDNHIIIKIAMDMSRFQQRFMELGREFENG
LFCDLVSTDMEDGYFTYKLLYDAGRNRIGINEAIVEKGSMTLMKHIKWEFDKLPHMLISGGTGGGKTYFI
LTLVKSLVASGADVRILDPKNADLADLEEVLEGKVFSRKNGIMMTLRKSVEDMMRRMDEMKNHPNYRTGE
NYAFLGYKPVFIVFDEFVAFVDMLDFKERESVIQDIKQIVMLGRQAGFFLVAGAQRPDAKYFADGIRDQF
NFRISLGKMSETGYAMLFGDTDKKFVEKDIKGRGYAYAGTGNIMEFYSPLIPNGYDFIEEIRLAKGA
MPVFLLLMILFWRSHLWDVYEKGIEDTFENFTFNIPLIVKSLLVSIIVPVALILVWYFIFHQSYKKIYHR
QKIARMIFSNKFYESKSVRSKITEKTVQKISYFPKFYYRVKDNHIIIKIAMDMSRFQQRFMELGREFENG
LFCDLVSTDMEDGYFTYKLLYDAGRNRIGINEAIVEKGSMTLMKHIKWEFDKLPHMLISGGTGGGKTYFI
LTLVKSLVASGADVRILDPKNADLADLEEVLEGKVFSRKNGIMMTLRKSVEDMMRRMDEMKNHPNYRTGE
NYAFLGYKPVFIVFDEFVAFVDMLDFKERESVIQDIKQIVMLGRQAGFFLVAGAQRPDAKYFADGIRDQF
NFRISLGKMSETGYAMLFGDTDKKFVEKDIKGRGYAYAGTGNIMEFYSPLIPNGYDFIEEIRLAKGA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 591669..615545
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| BPUM_RS02885 (BPUM_0549) | 588472..589590 | + | 1119 | WP_012009094 | AimR family lysis-lysogeny pheromone receptor | - |
| BPUM_RS19080 | 589587..589724 | + | 138 | WP_158511373 | hypothetical protein | - |
| BPUM_RS02890 (BPUM_0550) | 589910..590296 | - | 387 | WP_012009095 | helix-turn-helix transcriptional regulator | - |
| BPUM_RS02900 (BPUM_0552) | 590737..590979 | + | 243 | WP_012009096 | hypothetical protein | - |
| BPUM_RS19280 | 591038..591211 | + | 174 | WP_225970053 | hypothetical protein | - |
| BPUM_RS02905 (BPUM_03955) | 591669..592052 | + | 384 | WP_041815229 | YdcP family protein | orf23 |
| BPUM_RS02910 (BPUM_0553) | 592338..593294 | + | 957 | WP_041815231 | hypothetical protein | - |
| BPUM_RS02915 (BPUM_0554) | 593409..594662 | + | 1254 | WP_275477915 | FtsK/SpoIIIE domain-containing protein | virb4 |
| BPUM_RS02920 (BPUM_0555) | 594736..595836 | + | 1101 | WP_012009099 | replication initiation factor domain-containing protein | - |
| BPUM_RS02925 (BPUM_0556) | 596092..596349 | + | 258 | WP_012009100 | ASCH/PUA domain-containing protein | - |
| BPUM_RS02930 (BPUM_0557) | 596356..596634 | + | 279 | WP_012009101 | hypothetical protein | - |
| BPUM_RS02935 (BPUM_0558) | 596713..597363 | + | 651 | WP_012009102 | hypothetical protein | - |
| BPUM_RS02940 (BPUM_0559) | 597427..599967 | + | 2541 | WP_225970054 | thioester domain-containing protein | - |
| BPUM_RS02945 (BPUM_0560) | 600356..601180 | - | 825 | WP_012009104 | hypothetical protein | - |
| BPUM_RS02950 (BPUM_0561) | 601409..602194 | + | 786 | WP_012009105 | DNA cytosine methyltransferase | - |
| BPUM_RS02955 (BPUM_0562) | 602266..602562 | + | 297 | WP_012009106 | hypothetical protein | - |
| BPUM_RS02960 (BPUM_0563) | 602552..602833 | + | 282 | WP_012009107 | hypothetical protein | - |
| BPUM_RS02965 (BPUM_0564) | 602862..603254 | + | 393 | WP_012009108 | hypothetical protein | - |
| BPUM_RS02970 (BPUM_0565) | 603251..604480 | + | 1230 | WP_012009109 | CpaF/VirB11 family protein | virB11 |
| BPUM_RS02975 (BPUM_0566) | 604467..605279 | + | 813 | WP_012009110 | hypothetical protein | - |
| BPUM_RS02980 (BPUM_0567) | 605284..606045 | + | 762 | WP_012009111 | hypothetical protein | - |
| BPUM_RS02985 (BPUM_0568) | 606052..606411 | + | 360 | WP_012009112 | hypothetical protein | - |
| BPUM_RS19085 | 606425..606571 | + | 147 | WP_157840134 | hypothetical protein | - |
| BPUM_RS19090 (BPUM_0569) | 606593..606751 | + | 159 | WP_157840133 | hypothetical protein | - |
| BPUM_RS02995 (BPUM_0571) | 607239..608234 | + | 996 | WP_012009115 | conjugal transfer protein | orf13 |
| BPUM_RS03000 (BPUM_0572) | 608246..608506 | + | 261 | WP_012009116 | hypothetical protein | - |
| BPUM_RS03005 (BPUM_0573) | 608518..608985 | + | 468 | WP_012009117 | conjugal transfer protein | orf17b |
| BPUM_RS03010 (BPUM_0574) | 608963..611419 | + | 2457 | WP_012009118 | ATP-binding protein | virb4 |
| BPUM_RS03015 (BPUM_0575) | 612190..614571 | + | 2382 | WP_012009119 | hypothetical protein | orf15 |
| BPUM_RS03020 (BPUM_0576) | 614568..615545 | + | 978 | WP_012009120 | bifunctional lytic transglycosylase/C40 family peptidase | orf14 |
| BPUM_RS03025 (BPUM_0577) | 615555..616046 | + | 492 | WP_012009121 | hypothetical protein | - |
| BPUM_RS03030 (BPUM_0578) | 616074..616283 | + | 210 | WP_012009122 | DUF3173 domain-containing protein | - |
| BPUM_RS03035 (BPUM_0579) | 616280..616492 | + | 213 | WP_041815235 | hypothetical protein | - |
| BPUM_RS03040 (BPUM_0580) | 616489..617766 | + | 1278 | WP_012009123 | tyrosine-type recombinase/integrase | - |
| BPUM_RS03045 (BPUM_0581) | 618040..619098 | - | 1059 | WP_012009124 | AraC family transcriptional regulator | - |
Host bacterium
| ID | 441 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICEBpSAFR-032 | GenBank | NC_009848 |
| Element size | 3704641 bp | Coordinate of oriT [Strand] | 6282..6305 [+] |
| Host bacterium | Bacillus pumilus SAFR-032 | Coordinate of element | 588472..617766 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |