Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200495 |
| Name | oriT_ICEMlSym(R7A) |
| Organism | Mesorhizobium japonicum R7A |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | CP033366 (133586..133646 [+], 61 nt) |
| oriT length | 61 nt |
| IRs (inverted repeats) | 27..33, 37..43 (CCGCCTC..GAGGCGG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 61 nt
>oriT_ICEMlSym(R7A)
AAAACGATGTGCAGACAAAGGCTTCGCCGCCTCATGGAGGCGGTGCCTTTATCTTGCCTTA
AAAACGATGTGCAGACAAAGGCTTCGCCGCCTCATGGAGGCGGTGCCTTTATCTTGCCTTA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file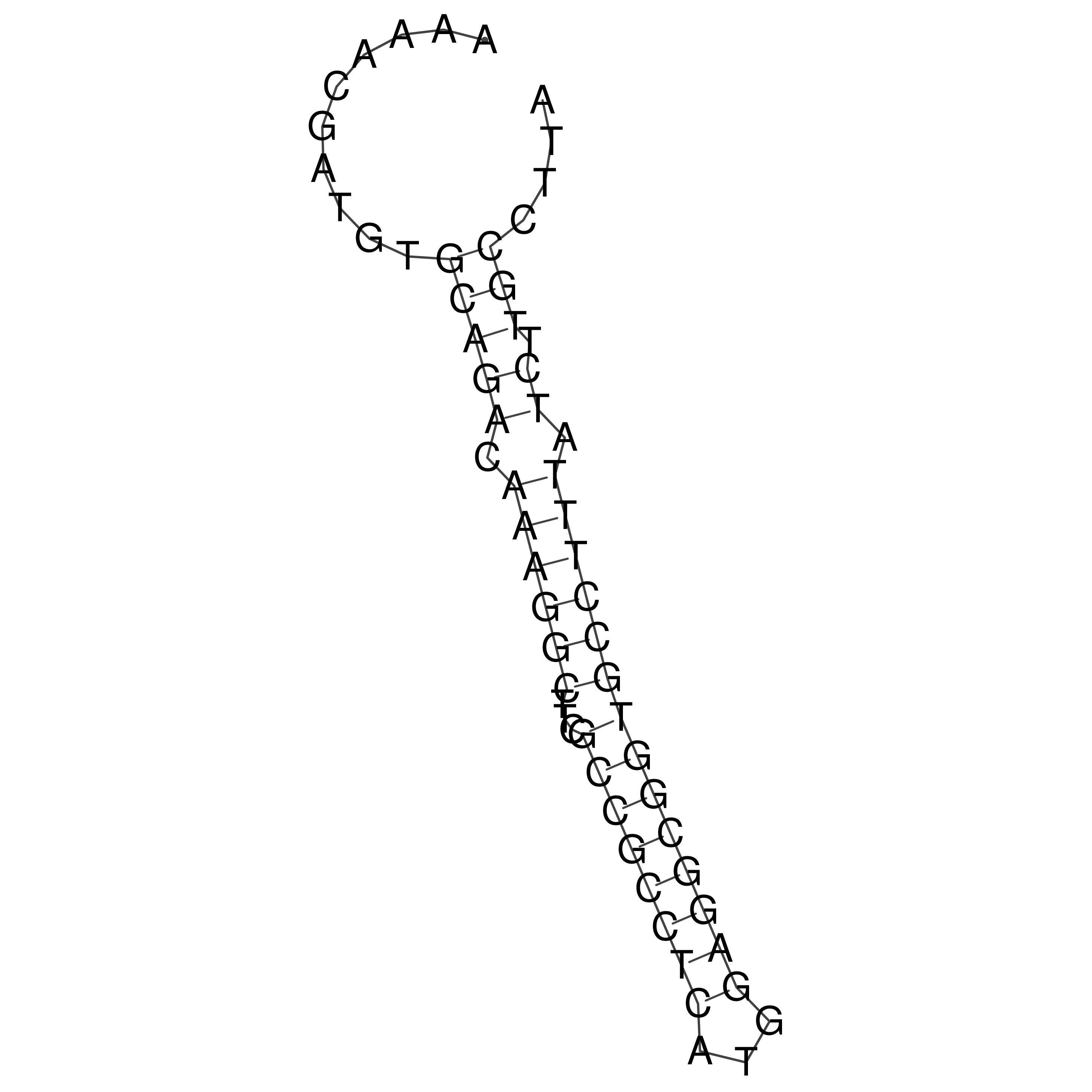
T4CP
| ID | 17152 | GenBank | QGX80473 |
| Name | t4cp2_EB234_29230_ICEMlSym(R7A) |
UniProt ID | _ |
| Length | 816 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 816 a.a. Molecular weight: 90272.73 Da Isoelectric Point: 5.9576
>QGX80473.1 conjugal transfer protein TrbE [Mesorhizobium japonicum R7A]
MLNLSEYRGRADRLADHLPWAALVAPGIVLNKDGSVQRTLRFRGPDLESSTEAELLGICARANNALRRLG
SGWTLFFEAERIEALGYPSSRFPDAASWLVDEERRAAFEGKVAHFESRYHLTLLSMPPPDAQARAESALV
DSHHSEGERDWRQEMARFRDETDRVLDLLSGFMPEVRPLDDAETLTYLHDTISTRRHPVAVPETPMYLDG
ILVDVPLTGGLEPMLGEQHLRTLTILGFPNLTRPGILDALNHQDFAYRWMTRFIPLDKTEATKTLTRLRR
QWFAKRKSIVAILREVVSNEPVPLVDSDADNKALDADEALQALGGDHVGFGYLTATVTVWDEDRQAAAEK
LRAVERIINGLGFTTIRESVNAVEAWLGSLPGHVYANVRQPLVHTLNLAHLMPLSSVWAGPAANEHLAEV
TGREAPPLLIAETSGSTPFRLSSHVEDVGHMLIVGPTGAGKSVLLALIALQFRRYAGAQVYVFDKGNSAR
AATLAMGGEHHVLGSDGALAFQPLRNIGDQGRRSWAAEWIAGLIAHENVTLTPEVKEAIWSALNSLATAP
AQERTLTGLSVLLQSNALKSALMPYTLDGPFGRLLDADDDRLALSDVQCFETEELMHSQSAVLPVLTYLF
HRLEERFDGRPTLLMLDEAWVYLDNPLFAARIREWLKVLRKKNVSVIFATQSLADIAGSAIAPAIIESCP
QRIFLPNDRAVEPQARTAYERFGLNNRQIELIARATPKRHYYLQSRCGNRLFELGLGPIALALCGASGPA
AQSLIDRVLSEHGQDSFVFRFLGARGLDWGAELLQQFPQPTKEQSS
MLNLSEYRGRADRLADHLPWAALVAPGIVLNKDGSVQRTLRFRGPDLESSTEAELLGICARANNALRRLG
SGWTLFFEAERIEALGYPSSRFPDAASWLVDEERRAAFEGKVAHFESRYHLTLLSMPPPDAQARAESALV
DSHHSEGERDWRQEMARFRDETDRVLDLLSGFMPEVRPLDDAETLTYLHDTISTRRHPVAVPETPMYLDG
ILVDVPLTGGLEPMLGEQHLRTLTILGFPNLTRPGILDALNHQDFAYRWMTRFIPLDKTEATKTLTRLRR
QWFAKRKSIVAILREVVSNEPVPLVDSDADNKALDADEALQALGGDHVGFGYLTATVTVWDEDRQAAAEK
LRAVERIINGLGFTTIRESVNAVEAWLGSLPGHVYANVRQPLVHTLNLAHLMPLSSVWAGPAANEHLAEV
TGREAPPLLIAETSGSTPFRLSSHVEDVGHMLIVGPTGAGKSVLLALIALQFRRYAGAQVYVFDKGNSAR
AATLAMGGEHHVLGSDGALAFQPLRNIGDQGRRSWAAEWIAGLIAHENVTLTPEVKEAIWSALNSLATAP
AQERTLTGLSVLLQSNALKSALMPYTLDGPFGRLLDADDDRLALSDVQCFETEELMHSQSAVLPVLTYLF
HRLEERFDGRPTLLMLDEAWVYLDNPLFAARIREWLKVLRKKNVSVIFATQSLADIAGSAIAPAIIESCP
QRIFLPNDRAVEPQARTAYERFGLNNRQIELIARATPKRHYYLQSRCGNRLFELGLGPIALALCGASGPA
AQSLIDRVLSEHGQDSFVFRFLGARGLDWGAELLQQFPQPTKEQSS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 17153 | GenBank | QGX80478 |
| Name | t4cp2_EB234_29255_ICEMlSym(R7A) |
UniProt ID | _ |
| Length | 676 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 676 a.a. Molecular weight: 75089.54 Da Isoelectric Point: 6.6518
>QGX80478.1 conjugal transfer protein TraG [Mesorhizobium japonicum R7A]
MTPTKLLIGQIAIVFAIVLLGVWAATQWCAHMLAFQEQLGAPWFVAAGWPIYEPWKLLEWWFQFDAYAPE
VFDKAGMLAGTSGFMGCAAAIAGSLWRARQRGLVTTYGSSRWAATREIEKAGLFQPAGVFLGKLKDRYLR
HDGPEHVMAFAPTRSGKGVGLVVPTLLSWSGSAVIHDIKGENWQLSSGWRSRFSHCLLFNPTDPRSARYN
PLLEVRKGSDEIRDVQNIADILVDPEGALERRNHWEKTSHSLLVGAILHVLYAEEEKTLARVATFLSDPQ
RSFAATLRRMMTTNHLGAANKPQVHPVVASAAREVLNKSENERSGVLSTAMSFLGLYRDPTVAAVTSDCD
WRIADLMDAEWPMSLYLVVPPSDISRTKPLVRLILNQIGRRLTERLEGDPKKSRKHQLLMMLDEFPALGR
LDFFETALAFMAGYGIRAYLIAQSLNQISKAYGDNNAILDNCHLRIAFSSNDERTAKRISDALGTATELR
SMRNYAGHRLAPWLSHVMVSRQETARPLLTPGEVMQLPPADELVLVSGLPPIRAKKLRYYEDHNFTERVL
PSPVLRDGPYADCPMSRPDDWTGQVRGVDRRLASDDESAGAALADEGGVQQQRHPGLPEEHTAVTQEPDQ
DDLSRPADDDSEAMADKRVMDRISTVAGAYGINEGRGDDKDIVPGF
MTPTKLLIGQIAIVFAIVLLGVWAATQWCAHMLAFQEQLGAPWFVAAGWPIYEPWKLLEWWFQFDAYAPE
VFDKAGMLAGTSGFMGCAAAIAGSLWRARQRGLVTTYGSSRWAATREIEKAGLFQPAGVFLGKLKDRYLR
HDGPEHVMAFAPTRSGKGVGLVVPTLLSWSGSAVIHDIKGENWQLSSGWRSRFSHCLLFNPTDPRSARYN
PLLEVRKGSDEIRDVQNIADILVDPEGALERRNHWEKTSHSLLVGAILHVLYAEEEKTLARVATFLSDPQ
RSFAATLRRMMTTNHLGAANKPQVHPVVASAAREVLNKSENERSGVLSTAMSFLGLYRDPTVAAVTSDCD
WRIADLMDAEWPMSLYLVVPPSDISRTKPLVRLILNQIGRRLTERLEGDPKKSRKHQLLMMLDEFPALGR
LDFFETALAFMAGYGIRAYLIAQSLNQISKAYGDNNAILDNCHLRIAFSSNDERTAKRISDALGTATELR
SMRNYAGHRLAPWLSHVMVSRQETARPLLTPGEVMQLPPADELVLVSGLPPIRAKKLRYYEDHNFTERVL
PSPVLRDGPYADCPMSRPDDWTGQVRGVDRRLASDDESAGAALADEGGVQQQRHPGLPEEHTAVTQEPDQ
DDLSRPADDDSEAMADKRVMDRISTVAGAYGINEGRGDDKDIVPGF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 17154 | GenBank | QGX80502 |
| Name | t4cp2_EB234_29415_ICEMlSym(R7A) |
UniProt ID | _ |
| Length | 581 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 581 a.a. Molecular weight: 64292.71 Da Isoelectric Point: 8.2057
>QGX80502.1 type IV secretion system protein VirD4 [Mesorhizobium japonicum R7A]
MSSTKTTSPNIAISIACSLALGFCAASLYATFRHGGSGEALMAFDVRAFWFETPFYLGFATPVFYQGAAI
VLSTSAVVLLIQQVVLRRKLEHHGTARWARVDEMRRTGYLRRYRGVTGPVFGKTSGPFWPGYYLTNGEQP
HSLIVAPTRAGKGVGIVIPTLLTFKGSVIALDVKGELFELTSRARKAAGHEVFKFAPLDSERRTNCYNPL
LDLIALHPQQQFTEARRLAANLITAKGEGAEGFVAGARDIFVAGILACIERGTPTIGAVYDLFAQPGEKF
KLFAQLAMETRNKEAQRIFDDMAGNDTKILTSYTSVLGDGGLNLWADPAVKAATSRSDFSVYDLRRRRTC
IYLCVGPNDLEVIAPLIRLFFQQIVSILQRSLPRQDEKHEVLFLLDEFKHLGKLEAIETAITTIAGYKGR
FMLIIQSLSALTGAYGEAGKQNFLSNTGLQVFMATADDETPNYISKAIGEYTFEAKSISYSQSRAFDRNI
QNSFQGAPLLRPEQVRLLNDEYEIVLIKGLPPLQLRKVRYFSDRILKRIFESQTGALPEPAPLTEAEEGF
SAESRLDEPLRGLAQDFDDTV
MSSTKTTSPNIAISIACSLALGFCAASLYATFRHGGSGEALMAFDVRAFWFETPFYLGFATPVFYQGAAI
VLSTSAVVLLIQQVVLRRKLEHHGTARWARVDEMRRTGYLRRYRGVTGPVFGKTSGPFWPGYYLTNGEQP
HSLIVAPTRAGKGVGIVIPTLLTFKGSVIALDVKGELFELTSRARKAAGHEVFKFAPLDSERRTNCYNPL
LDLIALHPQQQFTEARRLAANLITAKGEGAEGFVAGARDIFVAGILACIERGTPTIGAVYDLFAQPGEKF
KLFAQLAMETRNKEAQRIFDDMAGNDTKILTSYTSVLGDGGLNLWADPAVKAATSRSDFSVYDLRRRRTC
IYLCVGPNDLEVIAPLIRLFFQQIVSILQRSLPRQDEKHEVLFLLDEFKHLGKLEAIETAITTIAGYKGR
FMLIIQSLSALTGAYGEAGKQNFLSNTGLQVFMATADDETPNYISKAIGEYTFEAKSISYSQSRAFDRNI
QNSFQGAPLLRPEQVRLLNDEYEIVLIKGLPPLQLRKVRYFSDRILKRIFESQTGALPEPAPLTEAEEGF
SAESRLDEPLRGLAQDFDDTV
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 5922710..5931767
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| EB234_29185 | 5919081..5919620 | + | 540 | QGX80465 | hemerythrin domain-containing protein | - |
| EB234_29190 | 5919624..5920379 | - | 756 | QGX80466 | Crp/Fnr family transcriptional regulator | - |
| EB234_29195 | 5920537..5921886 | + | 1350 | QGX80467 | oxygen-independent coproporphyrinogen III oxidase | - |
| EB234_29200 | 5922425..5922697 | - | 273 | QGX80468 | DUF2274 domain-containing protein | - |
| EB234_29205 | 5922710..5923924 | - | 1215 | QGX81544 | TrbI/VirB10 family protein | virB10 |
| EB234_29210 | 5923936..5924964 | - | 1029 | QGX80469 | P-type conjugative transfer protein TrbG | virB9 |
| EB234_29215 | 5924961..5925686 | - | 726 | QGX80470 | conjugal transfer protein TrbF | virB8 |
| EB234_29220 | 5925687..5927009 | - | 1323 | QGX80471 | P-type conjugative transfer protein TrbL | virB6 |
| EB234_29225 | 5927041..5927766 | - | 726 | QGX80472 | P-type conjugative transfer protein TrbJ | virB5 |
| EB234_29230 | 5927763..5930213 | - | 2451 | QGX80473 | conjugal transfer protein TrbE | virb4 |
| EB234_29235 | 5930207..5930479 | - | 273 | QGX80474 | conjugal transfer protein | virB3 |
| EB234_29240 | 5930479..5930787 | - | 309 | QGX80475 | conjugal transfer protein TrbC | virB2 |
| EB234_29245 | 5930799..5931767 | - | 969 | QGX80476 | P-type conjugative transfer ATPase TrbB | virB11 |
| EB234_29250 | 5931764..5932177 | - | 414 | QGX80477 | CopG family transcriptional regulator | - |
| EB234_29255 | 5932296..5934326 | - | 2031 | QGX80478 | conjugal transfer protein TraG | - |
| EB234_29260 | 5934486..5934693 | + | 208 | Protein_5788 | XRE family transcriptional regulator | - |
| EB234_29265 | 5934988..5935377 | - | 390 | QGX80479 | hypothetical protein | - |
| EB234_29270 | 5935588..5936355 | + | 768 | QGX80480 | ZinT family metal-binding protein | - |
Region 2: 5952771..5962194
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| EB234_29325 | 5948375..5948502 | + | 128 | Protein_5801 | IS630 family transposase | - |
| EB234_29330 | 5948502..5949126 | + | 625 | Protein_5802 | ISKra4 family transposase | - |
| EB234_29335 | 5950108..5952648 | + | 2541 | QGX81546 | two-component system VirA-like sensor kinase | - |
| EB234_29340 | 5952771..5953811 | - | 1041 | QGX80488 | P-type DNA transfer ATPase VirB11 | virB11 |
| EB234_29345 | 5953795..5954934 | - | 1140 | QGX80489 | type IV secretion system protein VirB10 | virB10 |
| EB234_29350 | 5954931..5955812 | - | 882 | QGX80490 | type IV secretion system protein VirB9 | virB9 |
| EB234_29355 | 5955809..5956522 | - | 714 | QGX80491 | type IV secretion system protein VirB8 | virB8 |
| EB234_29360 | 5956519..5956677 | - | 159 | QGX80492 | type IV secretion system lipoprotein VirB7 | - |
| EB234_29365 | 5956704..5957591 | - | 888 | QGX80493 | type IV secretion system protein | virB6 |
| EB234_29370 | 5957649..5958323 | - | 675 | QGX80494 | type IV secretion system protein VirB5 | - |
| EB234_29375 | 5958337..5960706 | - | 2370 | QGX80495 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| EB234_29380 | 5960706..5961032 | - | 327 | QGX80496 | type IV secretion system protein VirB3 | virB3 |
| EB234_29385 | 5961032..5961397 | - | 366 | QGX80497 | pilin major subunit VirB2 | virB2 |
| EB234_29390 | 5961412..5962194 | - | 783 | QGX80498 | type IV secretion system lytic transglycosylase VirB1 | virB1 |
| EB234_29395 | 5963499..5964224 | + | 726 | QGX80499 | response regulator | - |
| EB234_29400 | 5964200..5964544 | - | 345 | QGX80500 | transposase domain-containing protein | - |
| EB234_29405 | 5964483..5964578 | - | 96 | Protein_5817 | hypothetical protein | - |
Host bacterium
| ID | 440 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICEMlSym(R7A) | GenBank | CP033366 |
| Element size | 6529994 bp | Coordinate of oriT [Strand] | 133586..133646 [+] |
| Host bacterium | Mesorhizobium japonicum R7A | Coordinate of element | 5899545..6401363 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | gmd |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | fixS, fixI, fixH, fixG, fixP, fixQ, fixO, fixN, nodM, nodD, nolL, nodB, eB234_29755, nodJ, nodI, nodC, nodA, nodS, mlr6097, nifU, nifX, nifN, nifE, nifK, nifD, nifH, nifQ, mLTONO_5203, nifS, nifW, fixA, fixB, fixC, fixX, nifA, nifB, nifZ, nifT, nolK, noeL, nodZ |
| Anti-CRISPR | AcrIIC1 |