Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200490 |
| Name | oriT_ICE_SsaL60_rpsI |
| Organism | Streptococcus salivarius integrative and |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | LT622833 (18628..18776 [+], 149 nt) |
| oriT length | 149 nt |
| IRs (inverted repeats) | 111..116, 124..129 (CACTTT..AAAGTG) 102..107, 111..116 (AAAGTG..CACTTT) 9..15, 28..34 (AACCCCC..GGGGGTT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 149 nt
>oriT_ICE_SsaL60_rpsI
GAGACTTCAACCCCCGATTTCTAATAGGGGGGTTACATTTGGCCAAAGTGCCACGTCCACCCCTCCTCATTCCTTGTGGGAATTGGGATTTCAGGATTTAGAAAGTGTGTCACTTTGGTCCAAAAAGTGTGTCACTTAGTCAAAAGGAG
GAGACTTCAACCCCCGATTTCTAATAGGGGGGTTACATTTGGCCAAAGTGCCACGTCCACCCCTCCTCATTCCTTGTGGGAATTGGGATTTCAGGATTTAGAAAGTGTGTCACTTTGGTCCAAAAAGTGTGTCACTTAGTCAAAAGGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file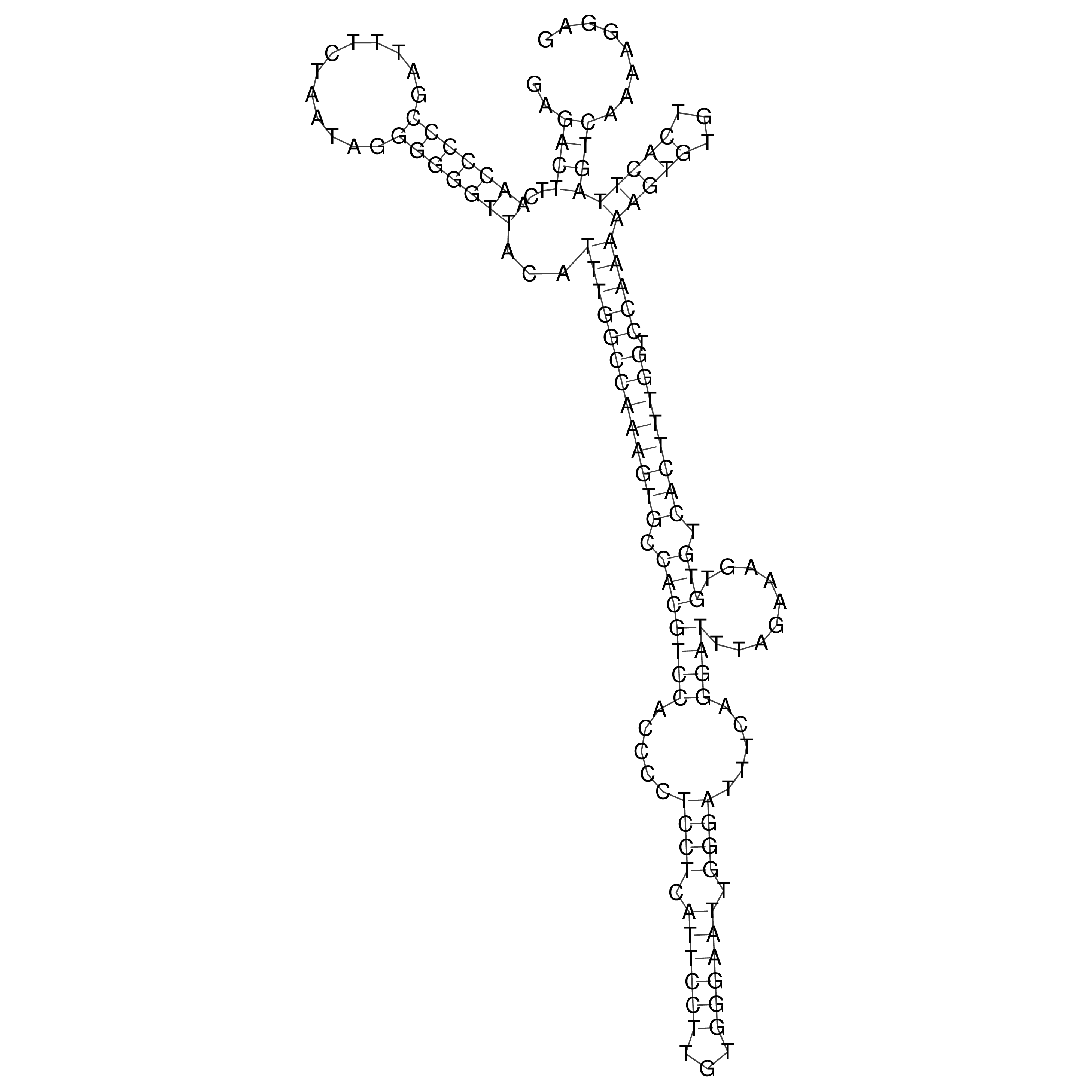
Relaxase
| ID | 14531 | GenBank | SCW20884 |
| Name | ORFJ_-_ICE_SsaL60_rpsI |
UniProt ID | _ |
| Length | 410 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 410 a.a. Molecular weight: 48329.77 Da Isoelectric Point: 6.0335
>SCW20884.1 putative relaxase [Streptococcus salivarius]
MTKMSPFQIKNFRKQTGLSQKAFAQAVNLPIRTYRSYESGERGLTIDKFRELKEKLGYYQECHRNSLRAH
IDYLRLTFPSLRDLEAFCENFLFCHLSEFTDQETRLMNYTHLWQRGNIWIFDFFDKSATNNYQTCLQLSG
QGCREMELLLEHKGISWQTFLQNILYAYEDVRVKRLDIALDELYKGYGHEDEHIQIPKLIDKLYAKEIVL
DTIRKWNITGGGSFTDNEDMEANHGLSLYFGSRQSQLYFNFYEKRYEIARMENISLEESLEIFGIWNRYE
LRFSDQKAQGVVEEYINGVDLGEIARGIVNKEIQVYDGVTRFGAYKPDEKWQELFGGVEPLKLSTSPQPY
SIERTIRWLTYQVANSLALVSEADKIMQTEYMKMIQNSGEITDRGEAILRLLKTNKHYEH
MTKMSPFQIKNFRKQTGLSQKAFAQAVNLPIRTYRSYESGERGLTIDKFRELKEKLGYYQECHRNSLRAH
IDYLRLTFPSLRDLEAFCENFLFCHLSEFTDQETRLMNYTHLWQRGNIWIFDFFDKSATNNYQTCLQLSG
QGCREMELLLEHKGISWQTFLQNILYAYEDVRVKRLDIALDELYKGYGHEDEHIQIPKLIDKLYAKEIVL
DTIRKWNITGGGSFTDNEDMEANHGLSLYFGSRQSQLYFNFYEKRYEIARMENISLEESLEIFGIWNRYE
LRFSDQKAQGVVEEYINGVDLGEIARGIVNKEIQVYDGVTRFGAYKPDEKWQELFGGVEPLKLSTSPQPY
SIERTIRWLTYQVANSLALVSEADKIMQTEYMKMIQNSGEITDRGEAILRLLKTNKHYEH
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 17147 | GenBank | SCW20883 |
| Name | ORFK_-_ICE_SsaL60_rpsI |
UniProt ID | _ |
| Length | 562 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 562 a.a. Molecular weight: 64908.99 Da Isoelectric Point: 7.2222
>SCW20883.1 FtsK/SpoIIIE family coupling protein [Streptococcus salivarius]
MKLFTYRGIRVRAFYRHIRGITWFIWWFPSLLAIGNYCYVNLEELKDHPLPQVYYIGGILVASCLVAYLL
NRMCYEKIVFFQKLNSLRILSRFLIENNLYLIKNIRRENKTIEKIVLPQVYLKQSRYKIEVRFILEGNKF
QDRFLNLGSTLEVMFNGDFRNKTFDKRFITYDIAINRIDSRITINEVEVKGSKLQLMTDVTWDYIEEPHL
LIGGGTGGGKTVILMTIIYALAKIGFVDICDPKNSDLAGLKRIPVFHGRVYTSKEDIIQCFKENVAFMEK
RYELMSSSPKFQAGKNFTHYGMTPKFVLVDEWAALMAKIDRDYSLQSELMEYLSQLVLEGRQAGVFIIFA
MQRPDGEFIKTALRDNFMKRLSVGHLESTGYDMMFGDANKTKEFKKLDEINGVKVKGRGYIANNGDLAGE
FFSPYVPLDQGFSFYDAYSKIPIMEFNGKEFEVFGESHQQSEPIEPIEEEKAPNTDTNLRPLKEFAEEQD
LKMATLRKIIYLLQEQGVGFERTESAILVNPFQEELLLESLIHFEEGGRKSYPKAVEATLSHHGLGQGQG
QA
MKLFTYRGIRVRAFYRHIRGITWFIWWFPSLLAIGNYCYVNLEELKDHPLPQVYYIGGILVASCLVAYLL
NRMCYEKIVFFQKLNSLRILSRFLIENNLYLIKNIRRENKTIEKIVLPQVYLKQSRYKIEVRFILEGNKF
QDRFLNLGSTLEVMFNGDFRNKTFDKRFITYDIAINRIDSRITINEVEVKGSKLQLMTDVTWDYIEEPHL
LIGGGTGGGKTVILMTIIYALAKIGFVDICDPKNSDLAGLKRIPVFHGRVYTSKEDIIQCFKENVAFMEK
RYELMSSSPKFQAGKNFTHYGMTPKFVLVDEWAALMAKIDRDYSLQSELMEYLSQLVLEGRQAGVFIIFA
MQRPDGEFIKTALRDNFMKRLSVGHLESTGYDMMFGDANKTKEFKKLDEINGVKVKGRGYIANNGDLAGE
FFSPYVPLDQGFSFYDAYSKIPIMEFNGKEFEVFGESHQQSEPIEPIEEEKAPNTDTNLRPLKEFAEEQD
LKMATLRKIIYLLQEQGVGFERTESAILVNPFQEELLLESLIHFEEGGRKSYPKAVEATLSHHGLGQGQG
QA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 435 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICE_SsaL60_rpsI | GenBank | LT622833 |
| Element size | 31357 bp | Coordinate of oriT [Strand] | 18628..18776 [+] |
| Host bacterium | Streptococcus salivarius integrative and | Coordinate of element | 1..31357 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | cadX |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |