Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200475 |
| Name | oriT_ICECmeCH34-1 |
| Organism | Cupriavidus metallidurans CH34 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | CP000352 (8299..8616 [-], 318 nt) |
| oriT length | 318 nt |
| IRs (inverted repeats) | 238..243, 251..256 (CCAGCC..GGCTGG) 199..206, 215..222 (TAAAAGGC..GCCTTTTA) 197..202, 207..212 (TTTAAA..TTTAAA) |
| Location of nic site | 44..45 |
| Conserved sequence flanking the nic site |
TCGTGCCTGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 318 nt
>oriT_ICECmeCH34-1
CCATGGCCGATCGATCGTGCTTGCGCCAGCGAAGAAAGGTCGTGCCTGCGCCAGTGGTCTGCTGGGCCAGCTCGACGGGAAGCAGGTGGAAGGGATGCCCGCACGCCTGTGGCAGCACCTGTCGCTGCGCCAGGGCAATCAACTCGTCGCGCATGGCGAAGCACTGGCTGGCCCAGGCCTCGAAGTCCCCTTTACCTTTAAAAGGCTTTAAAAGGCCTTTTAGAGAGGCCGCGTGTTCCAGCCGCATGAAGGCTGGCTGTTCCAGACGACGGAAGTAGCGCGGTTCGCGGTTCGGGTCGCTCATGCCGGTTCGTCCTC
CCATGGCCGATCGATCGTGCTTGCGCCAGCGAAGAAAGGTCGTGCCTGCGCCAGTGGTCTGCTGGGCCAGCTCGACGGGAAGCAGGTGGAAGGGATGCCCGCACGCCTGTGGCAGCACCTGTCGCTGCGCCAGGGCAATCAACTCGTCGCGCATGGCGAAGCACTGGCTGGCCCAGGCCTCGAAGTCCCCTTTACCTTTAAAAGGCTTTAAAAGGCCTTTTAGAGAGGCCGCGTGTTCCAGCCGCATGAAGGCTGGCTGTTCCAGACGACGGAAGTAGCGCGGTTCGCGGTTCGGGTCGCTCATGCCGGTTCGTCCTC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file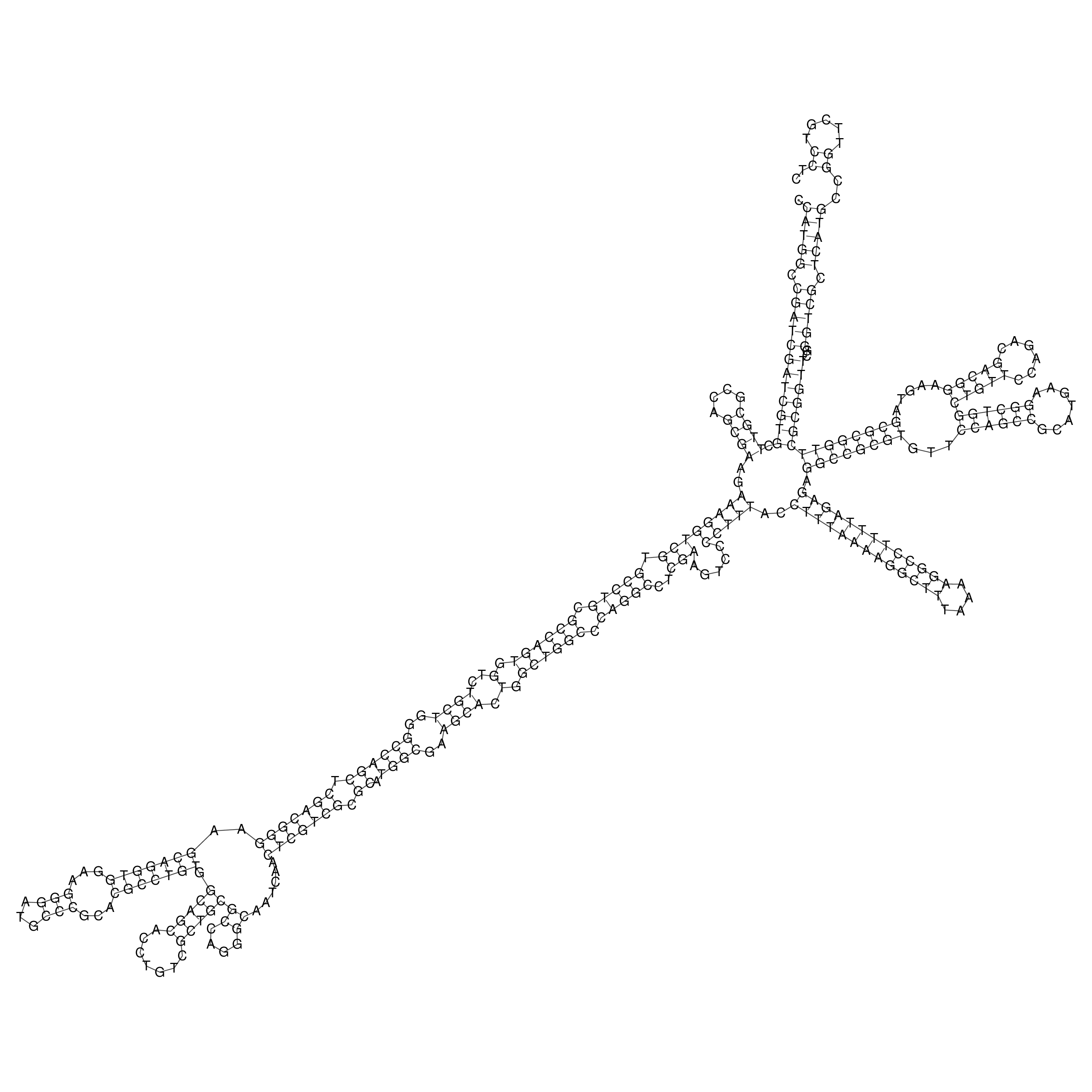
Relaxase
| ID | 14512 | GenBank | ABF09241 |
| Name | TraI_2_Rmet_2364_ICECmeCH34-1 |
UniProt ID | _ |
| Length | 680 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 680 a.a. Molecular weight: 74533.78 Da Isoelectric Point: 7.8320
>ABF09241.1 relaxase [Cupriavidus metallidurans CH34]
MTLESHPVPRYTETVKGQRAEMARERGARGKALPRKGQKASRSARHQDTHMLSLFQRKRPAVATAPTPPP
ASDLPKGLMRPESAASLLATPRRQKLLEHIWQRTSLSRKQFAVLYRAPLERYAELVQAFPASESHHHAYP
GGMLDHGLEIVAYALKLRQSHLLPIGASPEDQAAQAEAWTAAVAYAALLHDIGKIAVDLHVELADGSLWH
PWYGPLHQPYRFRYRDDREYRLHSAATGLLYRQLLDTQLLDWLSDYRDLWGPLLYVLAGQYEHAGVLGEL
VVQADRASVAQELGGDPARAMAAPKHALQRKLLDGLRYLLKEELKLNQPEASDGWLTEDALWLVSKTVSD
KLRAHLLSQGIDGIPANNTAVFNVLQDHGMLQPTLDGKAVWRATVTSNAGWTHSFTLLRLAPALIWEAGE
RPAPFAGTVAIDTSPVEKHAAAASSPEVAAKPTPGGQESPPWEGGSAADPIIAPLAEAMPDVMEDLLTMV
GMGDSSATQQDESTIPTELSATRPEAPPPSIVASSPSSPAPAATPAATTVQPSGEHFMAWLKQGIASRRL
IINDAKALVHTVSDTAYLVSPGVFQRYAQEHPQLAAIARQEKLEPWQWAQKRFEKLAAHRKQASGLNIWT
CHVSGPRKSRQLHGYLLTQPGALFERVPPNNPYLCLFNEEAGREVSLAAK
MTLESHPVPRYTETVKGQRAEMARERGARGKALPRKGQKASRSARHQDTHMLSLFQRKRPAVATAPTPPP
ASDLPKGLMRPESAASLLATPRRQKLLEHIWQRTSLSRKQFAVLYRAPLERYAELVQAFPASESHHHAYP
GGMLDHGLEIVAYALKLRQSHLLPIGASPEDQAAQAEAWTAAVAYAALLHDIGKIAVDLHVELADGSLWH
PWYGPLHQPYRFRYRDDREYRLHSAATGLLYRQLLDTQLLDWLSDYRDLWGPLLYVLAGQYEHAGVLGEL
VVQADRASVAQELGGDPARAMAAPKHALQRKLLDGLRYLLKEELKLNQPEASDGWLTEDALWLVSKTVSD
KLRAHLLSQGIDGIPANNTAVFNVLQDHGMLQPTLDGKAVWRATVTSNAGWTHSFTLLRLAPALIWEAGE
RPAPFAGTVAIDTSPVEKHAAAASSPEVAAKPTPGGQESPPWEGGSAADPIIAPLAEAMPDVMEDLLTMV
GMGDSSATQQDESTIPTELSATRPEAPPPSIVASSPSSPAPAATPAATTVQPSGEHFMAWLKQGIASRRL
IINDAKALVHTVSDTAYLVSPGVFQRYAQEHPQLAAIARQEKLEPWQWAQKRFEKLAAHRKQASGLNIWT
CHVSGPRKSRQLHGYLLTQPGALFERVPPNNPYLCLFNEEAGREVSLAAK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 17128 | GenBank | ABF09213 |
| Name | virD4_Rmet_2336_ICECmeCH34-1 |
UniProt ID | _ |
| Length | 729 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 729 a.a. Molecular weight: 80777.58 Da Isoelectric Point: 7.3460
>ABF09213.1 VirD4, Type IV secretory pathway, VirD4 components [Cupriavidus metallidurans CH34]
MSGKQPVEVLLRPAVELYTVAACAGAAFLCLVAPWSLALSPAMGIGSALAFGAYGAIRYRDARIILRYRR
NIRRLPRYVMTSKDVPVSQQRLFVGRGFLWEQKHTHRLMQTYRPEFRRYVEPTPAYRLTRRLEERLEFAP
LPLSRLPKLTGWDAPFNPVRPLPPVGGLPRLHGIEPDEVDVSLPLGERVGHSLVLGTTRVGKTRLAELFV
TQDIRRRNAAGEHEVVIVIDPKGDADLLKRMYVEAQRAGREGEFYIFHLGWPEISARYNAVGRFGRISEV
ATRIAGQLSGEGNSAAFREFAWRFVNIIARALVELGQRPDYMLIQRHVINIDALFIEYAQHYFAKTEPKA
WEVIVQIEAKLNEKNIPRNMIGREKRVVALEQYLSQARNYDPVLDGLRSAVRYDKTYFDKIVASLLPLLE
KLTSGKIAQLLAPNYSDLADPRPIFDWMQVIRKRAVVYVGLDALSDAEVAAAVGNSMFSDLVSVAGHIYK
HGIDDGLPGASAGARVPINVHADEFNELMGDEFIPLINKGGGAGLQVTAYTQTLSDIEARIGNRAKAGQV
IGNFNNLFMLRVRETATAELLTRQLPKVEVYTTTIVSGATDSSDIRGATDFTSNTQDRISMASVPMIEPS
HVVGLPKGQCFALLQGGQLWKVRMPLPAPDPDEVMPTDLQQLAGYMRQSYSEATQWWEFTSSAALPHAAL
PDDLLDDAAPAEPDAVATGADDSTGEAAP
MSGKQPVEVLLRPAVELYTVAACAGAAFLCLVAPWSLALSPAMGIGSALAFGAYGAIRYRDARIILRYRR
NIRRLPRYVMTSKDVPVSQQRLFVGRGFLWEQKHTHRLMQTYRPEFRRYVEPTPAYRLTRRLEERLEFAP
LPLSRLPKLTGWDAPFNPVRPLPPVGGLPRLHGIEPDEVDVSLPLGERVGHSLVLGTTRVGKTRLAELFV
TQDIRRRNAAGEHEVVIVIDPKGDADLLKRMYVEAQRAGREGEFYIFHLGWPEISARYNAVGRFGRISEV
ATRIAGQLSGEGNSAAFREFAWRFVNIIARALVELGQRPDYMLIQRHVINIDALFIEYAQHYFAKTEPKA
WEVIVQIEAKLNEKNIPRNMIGREKRVVALEQYLSQARNYDPVLDGLRSAVRYDKTYFDKIVASLLPLLE
KLTSGKIAQLLAPNYSDLADPRPIFDWMQVIRKRAVVYVGLDALSDAEVAAAVGNSMFSDLVSVAGHIYK
HGIDDGLPGASAGARVPINVHADEFNELMGDEFIPLINKGGGAGLQVTAYTQTLSDIEARIGNRAKAGQV
IGNFNNLFMLRVRETATAELLTRQLPKVEVYTTTIVSGATDSSDIRGATDFTSNTQDRISMASVPMIEPS
HVVGLPKGQCFALLQGGQLWKVRMPLPAPDPDEVMPTDLQQLAGYMRQSYSEATQWWEFTSSAALPHAAL
PDDLLDDAAPAEPDAVATGADDSTGEAAP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 2545874..2573458
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| Rmet_2327 | 2541444..2542553 | + | 1110 | ABF09204 | putative adenine-specific DNA methyltransferase | - |
| Rmet_2328 | 2542604..2542924 | + | 321 | ABF09205 | conserved hypothetical protein | - |
| Rmet_2329 | 2543014..2543319 | + | 306 | ABF09206 | conserved hypothetical protein | - |
| Rmet_2330 | 2543456..2545735 | + | 2280 | ABF09207 | Putative plasmid-related DNA/RNA helicase | - |
| Rmet_2331 | 2545874..2546473 | + | 600 | ABF09208 | pilL lipoprotein (type IV pili) | tfc2 |
| Rmet_2332 | 2546470..2547114 | + | 645 | ABF09209 | conserved putative exported protein | - |
| Rmet_2333 | 2547127..2547864 | + | 738 | ABF09210 | conserved putative exported protein | tfc3 |
| Rmet_2334 | 2547846..2548436 | + | 591 | ABF09211 | putative lytic transglycosylase, catalytic | virB1 |
| Rmet_2335 | 2548433..2548981 | + | 549 | ABF09212 | putative exported protein | tfc5 |
| Rmet_2336 | 2548986..2551175 | + | 2190 | ABF09213 | VirD4, Type IV secretory pathway, VirD4 components | - |
| Rmet_2337 | 2551172..2551921 | + | 750 | ABF09214 | conserved hypothetical protein; putative membrane protein | tfc7 |
| Rmet_2338 | 2551962..2554586 | - | 2625 | ABF09215 | conserved hypothetical protein | - |
| Rmet_2339 | 2554596..2555321 | - | 726 | ABF09216 | conserved hypothetical protein | - |
| Rmet_2340 | 2555360..2556262 | - | 903 | ABF09217 | conserved hypothetical protein | - |
| Rmet_2341 | 2556262..2556915 | - | 654 | ABF09218 | conserved hypothetical protein | - |
| Rmet_6523 | 2556759..2557229 | - | 471 | ADC45130 | conserved hypothetical protein | - |
| Rmet_2342 | 2557226..2558140 | - | 915 | ABF09219 | putative AAA ATPase, central region | - |
| Rmet_2343 | 2558158..2558733 | - | 576 | ABF09220 | conserved hypothetical protein | - |
| Rmet_2344 | 2558730..2559200 | - | 471 | ABF09221 | putative excisionase | - |
| Rmet_2345 | 2559404..2559787 | + | 384 | ABF09222 | conserved hypothetical protein | tfc8 |
| Rmet_2346 | 2559784..2560017 | + | 234 | ABF09223 | conserved hypothetical protein | tfc9 |
| Rmet_2347 | 2560034..2560393 | + | 360 | ABF09224 | conserved hypothetical protein | tfc10 |
| Rmet_2348 | 2560406..2560804 | + | 399 | ABF09225 | conserved hypothetical protein | tfc11 |
| Rmet_2349 | 2560801..2561493 | + | 693 | ABF09226 | conserved hypothetical protein | tfc12 |
| Rmet_2350 | 2561490..2562401 | + | 912 | ABF09227 | conserved hypothetical protein | tfc13 |
| Rmet_2351 | 2562391..2563809 | + | 1419 | ABF09228 | conserved hypothetical protein | tfc14 |
| Rmet_2352 | 2563790..2564230 | + | 441 | ABF09229 | Putative secreted protein | tfc15 |
| Rmet_2353 | 2564230..2567121 | + | 2892 | ABF09230 | Type IV secretory pathway, VirB4 components | virb4 |
| Rmet_2354 | 2567146..2567910 | + | 765 | ABF09231 | putative exported protein | - |
| Rmet_2355 | 2568086..2568580 | + | 495 | ABF09232 | DNA binding protein, possibly associated to lesions | - |
| Rmet_2356 | 2568745..2569191 | + | 447 | ABF09233 | conserved hypothetical protein; putative exported protein | tfc24 |
| Rmet_2357 | 2569188..2570138 | + | 951 | ABF09234 | putative exported protein | tfc23 |
| Rmet_2358 | 2570148..2571542 | + | 1395 | ABF09235 | putative membrane protein | tfc22 |
| Rmet_2359 | 2571539..2571895 | + | 357 | ABF09236 | putative membrane protein | tfc18 |
| Rmet_2360 | 2571911..2573458 | + | 1548 | ABF09237 | conserved hypothetical protein (membrane) | tfc19 |
| Rmet_2361 | 2573474..2573833 | - | 360 | ABF09238 | conserved hypothetical protein | - |
| Rmet_2362 | 2573907..2574539 | - | 633 | ABF09239 | conserved hypothetical protein | - |
| Rmet_2363 | 2574552..2575178 | - | 627 | ABF09240 | conserved hypothetical protein | - |
| Rmet_2364 | 2575344..2577386 | + | 2043 | ABF09241 | relaxase | - |
| Rmet_2365 | 2577434..2578429 | - | 996 | ABF09242 | putative transcriptional regulator | - |
Host bacterium
| ID | 420 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICECmeCH34-1 | GenBank | CP000352 |
| Element size | 3928089 bp | Coordinate of oriT [Strand] | 8299..8616 [-] |
| Host bacterium | Cupriavidus metallidurans CH34 | Coordinate of element | 2508632..2618212 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | pbrA, merR2, merT |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA3 |