Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200464 |
| Name | oriT_ICEBxyXB1A-1 |
| Organism | Bacteroides xylanisolvens XB1A draft |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | FP929033 (18837..19038 [+], 202 nt) |
| oriT length | 202 nt |
| IRs (inverted repeats) | 101..107, 120..126 (GGCTTCC..GGAAGCC) 13..19, 24..30 (TATGTTC..GAACATA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 202 nt
>oriT_ICEBxyXB1A-1
CCGACGGATTTTTATGTTCGTCTGAACATAGCAAGATGTCTTTTCAGCCGCTCAAAACGTTTTGAGCAGCCACGAAAAGCACTTGCCCTTGCAGGGGGCGGGCTTCCTCTCCGAAGTCGGGAAGCCTTTCAAACCTGCGGTGTCTGTGCCAAGAAGCGGCGGCTTATGCCGTCCATCCGCTAAATCCAAAGTTTATATGAAA
CCGACGGATTTTTATGTTCGTCTGAACATAGCAAGATGTCTTTTCAGCCGCTCAAAACGTTTTGAGCAGCCACGAAAAGCACTTGCCCTTGCAGGGGGCGGGCTTCCTCTCCGAAGTCGGGAAGCCTTTCAAACCTGCGGTGTCTGTGCCAAGAAGCGGCGGCTTATGCCGTCCATCCGCTAAATCCAAAGTTTATATGAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file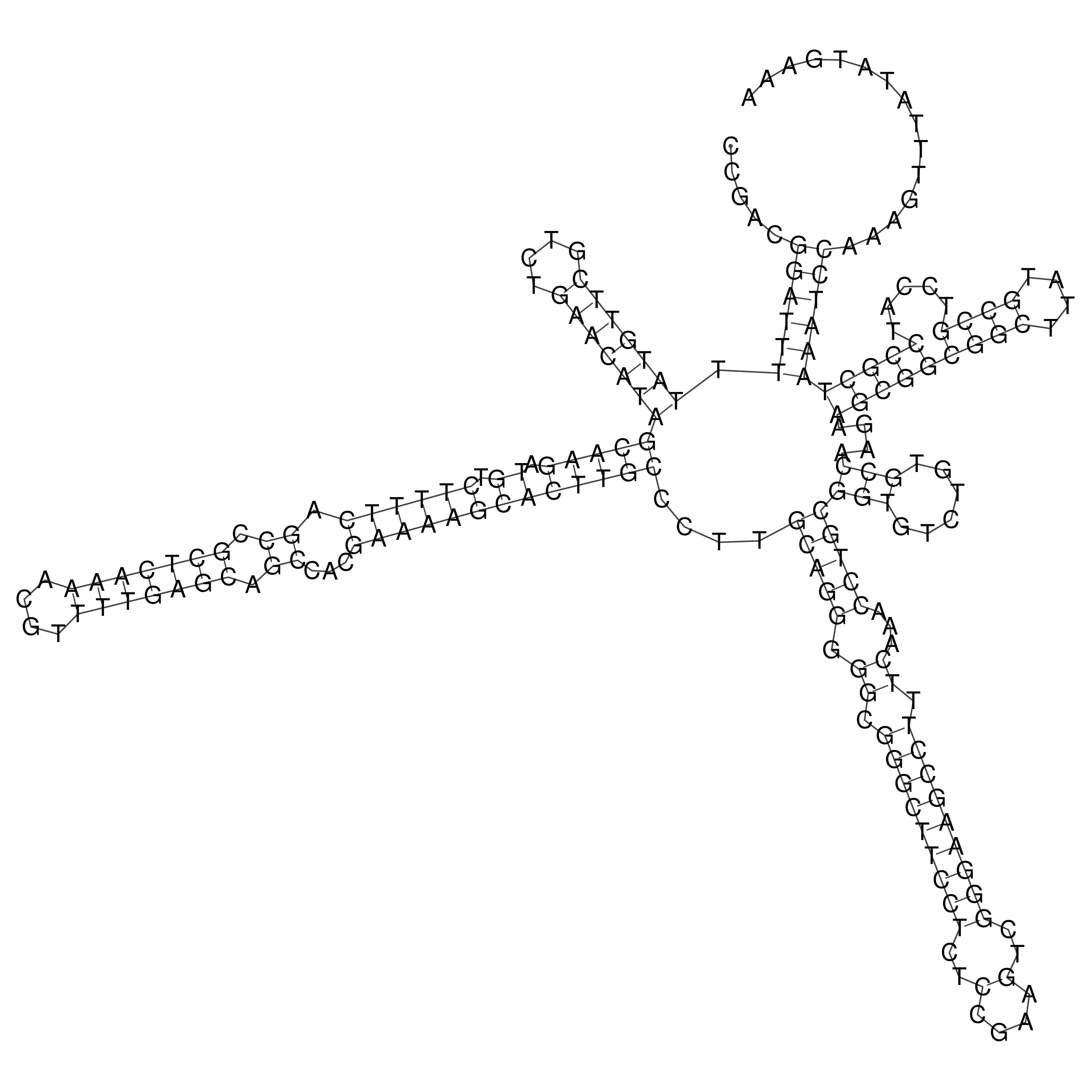
Relaxase
| ID | 14488 | GenBank | CBK68382 |
| Name | Relaxase_BXY_33850_ICEBxyXB1A-1 |
UniProt ID | _ |
| Length | 416 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 416 a.a. Molecular weight: 47047.49 Da Isoelectric Point: 9.8228
>CBK68382.1 Relaxase/Mobilisation nuclease domain [Bacteroides xylanisolvens XB1A]
MVAKISVGKSLYGALAYNGEKINEAKGRLLTTNRIYNDGTGTVDIRKAMEGFLACMPEHTRVEKPVLHIS
LNPHPDDVLTDTELQDIAREYLEKLGYGNQPYLVVKHEDIDRHHLHIVTINVDEKGRRLNQDFLFRRSDR
IRRELEQKYGLHPAERKNQRIENPLRKVDASAGDVKRQIGNTVKALNGQYRFQTMGEYRALLSLYNMTVE
EARGNVRGREYHGLVYSVTDDAGNKTGNPFKSSLFGKSAGYEAVQKKFARSKQEIKDRKLADMTKRAVLS
VLEGTYDKEKFVSRLREKGIDTVLRYTEEGRIYGATFIDHRTGCVLNGSRMGKELSANALQEHFTLPYAG
QPPIPLSVPVETGEDMRRQTASDHEDTVGGVGLLTPEGPAVDAEEEAFIRAMQRKKKKKRRKGLGM
MVAKISVGKSLYGALAYNGEKINEAKGRLLTTNRIYNDGTGTVDIRKAMEGFLACMPEHTRVEKPVLHIS
LNPHPDDVLTDTELQDIAREYLEKLGYGNQPYLVVKHEDIDRHHLHIVTINVDEKGRRLNQDFLFRRSDR
IRRELEQKYGLHPAERKNQRIENPLRKVDASAGDVKRQIGNTVKALNGQYRFQTMGEYRALLSLYNMTVE
EARGNVRGREYHGLVYSVTDDAGNKTGNPFKSSLFGKSAGYEAVQKKFARSKQEIKDRKLADMTKRAVLS
VLEGTYDKEKFVSRLREKGIDTVLRYTEEGRIYGATFIDHRTGCVLNGSRMGKELSANALQEHFTLPYAG
QPPIPLSVPVETGEDMRRQTASDHEDTVGGVGLLTPEGPAVDAEEEAFIRAMQRKKKKKRRKGLGM
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 17096 | GenBank | CBK68383 |
| Name | t4cp2_BXY_33860_ICEBxyXB1A-1 |
UniProt ID | _ |
| Length | 669 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 669 a.a. Molecular weight: 76691.71 Da Isoelectric Point: 7.1005
>CBK68383.1 Type IV secretory pathway, VirD4 components [Bacteroides xylanisolvens XB1A]
MSQQEDDLRALAKIMDFLRAVSIILVVMNVYWFCYEAIRLWGVDIGVVDRILMNFDRTAGLFRSILYTKL
FAVLLLALSCLGTKGVKGEKITWGRIWTALAAGFVLFFLNWWILALPLPVEAVTGLYVLTVGTGYVCLLM
GGLWMSRLLKHNLMDDVFNNENESFMQETRLIESEYSVNLPTRFYYRKRWNNGWINVVNPFRASIVLGTP
GSGKSYAVVNNFIKQQIEKGFSMYVYDFKFSDLSTIAYNHLLNHPEGYKVKPKFYVINFDDPRRSHRCNP
IHPDFMEDITDAYESAYTIMLNLNKSWVQKQGDFFVESPIILFAAIIWYLKIFQNGKYCTFPHAIEFLNR
RYEDIFPILTSYPELENYLSPFMDAWLGGAAEQLMGQIASAKIPLSRMISPQLYWVMSDSEFTLDINNPE
EPKILCVGNNPDRQNIYGAALGLYNSRIVKLINKKGMLKSSVIIDELPTIYFKGLDNLIATARSNKVAVC
LGFQDFSQLVRDYGDKEAKVVMNTVGNIFSGQVVGETAKTLSERFGKVLQKRQSISINRQDVSTSINTQM
DALIPPSKISGLTQGMFVGSVSDNFNERIEQKIFHCEIVVDAEKVKREEKAYKPIPIITDFTDEDGKDCM
KETVQANYRRIKEEVKQIVQEELERIANDENLKHLLQQK
MSQQEDDLRALAKIMDFLRAVSIILVVMNVYWFCYEAIRLWGVDIGVVDRILMNFDRTAGLFRSILYTKL
FAVLLLALSCLGTKGVKGEKITWGRIWTALAAGFVLFFLNWWILALPLPVEAVTGLYVLTVGTGYVCLLM
GGLWMSRLLKHNLMDDVFNNENESFMQETRLIESEYSVNLPTRFYYRKRWNNGWINVVNPFRASIVLGTP
GSGKSYAVVNNFIKQQIEKGFSMYVYDFKFSDLSTIAYNHLLNHPEGYKVKPKFYVINFDDPRRSHRCNP
IHPDFMEDITDAYESAYTIMLNLNKSWVQKQGDFFVESPIILFAAIIWYLKIFQNGKYCTFPHAIEFLNR
RYEDIFPILTSYPELENYLSPFMDAWLGGAAEQLMGQIASAKIPLSRMISPQLYWVMSDSEFTLDINNPE
EPKILCVGNNPDRQNIYGAALGLYNSRIVKLINKKGMLKSSVIIDELPTIYFKGLDNLIATARSNKVAVC
LGFQDFSQLVRDYGDKEAKVVMNTVGNIFSGQVVGETAKTLSERFGKVLQKRQSISINRQDVSTSINTQM
DALIPPSKISGLTQGMFVGSVSDNFNERIEQKIFHCEIVVDAEKVKREEKAYKPIPIITDFTDEDGKDCM
KETVQANYRRIKEEVKQIVQEELERIANDENLKHLLQQK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 4232472..4243046
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| BXY_33550 | 4227623..4228075 | + | 453 | CBK68356 | hypothetical protein | - |
| BXY_33560 | 4228087..4228332 | - | 246 | CBK68357 | hypothetical protein | - |
| BXY_33570 | 4228464..4228685 | + | 222 | CBK68358 | hypothetical protein | - |
| BXY_33580 | 4228699..4229121 | + | 423 | CBK68359 | hypothetical protein | - |
| BXY_33600 | 4229562..4230413 | + | 852 | CBK68360 | hypothetical protein | - |
| BXY_33610 | 4230438..4230695 | + | 258 | CBK68361 | hypothetical protein | - |
| BXY_33620 | 4230728..4230949 | + | 222 | CBK68362 | hypothetical protein | - |
| BXY_33630 | 4230946..4231191 | + | 246 | CBK68363 | hypothetical protein | - |
| BXY_33640 | 4231237..4231545 | + | 309 | CBK68364 | hypothetical protein | - |
| BXY_33650 | 4231663..4231884 | + | 222 | CBK68365 | hypothetical protein | - |
| BXY_33660 | 4231948..4232475 | - | 528 | CBK68366 | Phage-related lysozyme (muraminidase) | - |
| BXY_33670 | 4232472..4232975 | - | 504 | CBK68367 | hypothetical protein | traQ |
| BXY_33680 | 4232972..4233883 | - | 912 | CBK68368 | DNA primase (bacterial type) | traP |
| BXY_33690 | 4233883..4234377 | - | 495 | CBK68369 | Conjugative transposon protein TraO. | traO |
| BXY_33700 | 4234461..4235447 | - | 987 | CBK68370 | hypothetical protein | traN |
| BXY_33710 | 4235492..4236862 | - | 1371 | CBK68371 | Protein of unknown function (DUF3714). | traM |
| BXY_33720 | 4236831..4237151 | - | 321 | CBK68372 | hypothetical protein | traL |
| BXY_33730 | 4237158..4237781 | - | 624 | CBK68373 | hypothetical protein | traK |
| BXY_33740 | 4237813..4238817 | - | 1005 | CBK68374 | Homologues of TraJ from Bacteroides conjugative transposon. | traJ |
| BXY_33760 | 4239474..4239833 | - | 360 | CBK68375 | hypothetical protein | traH |
| BXY_33770 | 4239884..4242376 | - | 2493 | CBK68376 | Domain of unknown function DUF87. | virb4 |
| BXY_33790 | 4242735..4243046 | - | 312 | CBK68377 | hypothetical protein | traE |
| BXY_33800 | 4243249..4243977 | - | 729 | CBK68378 | hypothetical protein | - |
| BXY_33810 | 4243999..4244346 | - | 348 | CBK68379 | Protein of unknown function (DUF3408). | - |
| BXY_33820 | 4244349..4244789 | - | 441 | CBK68380 | Protein of unknown function (DUF3408). | - |
| BXY_33840 | 4246264..4246689 | + | 426 | CBK68381 | hypothetical protein | - |
| BXY_33850 | 4246668..4247918 | + | 1251 | CBK68382 | Relaxase/Mobilisation nuclease domain. | - |
Host bacterium
| ID | 409 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICEBxyXB1A-1 | GenBank | FP929033 |
| Element size | 5976145 bp | Coordinate of oriT [Strand] | 18837..19038 [+] |
| Host bacterium | Bacteroides xylanisolvens XB1A draft | Coordinate of element | 4227232..4279471 |
Cargo genes
| Drug resistance gene | tet(Q) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA9 |