Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200460 |
| Name | oriT1_ICESgal1 |
| Organism | Streptococcus gallolyticus UCN34 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | FN597254 (45728..45860 [-], 133 nt) |
| oriT length | 133 nt |
| IRs (inverted repeats) | 65..70, 83..88 (AAATCC..GGATTT) 6..12, 23..29 (ACCCCCC..GGGGGGT) |
| Location of nic site | 75..76 |
| Conserved sequence flanking the nic site |
TTTGGTTACA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 133 nt
ACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file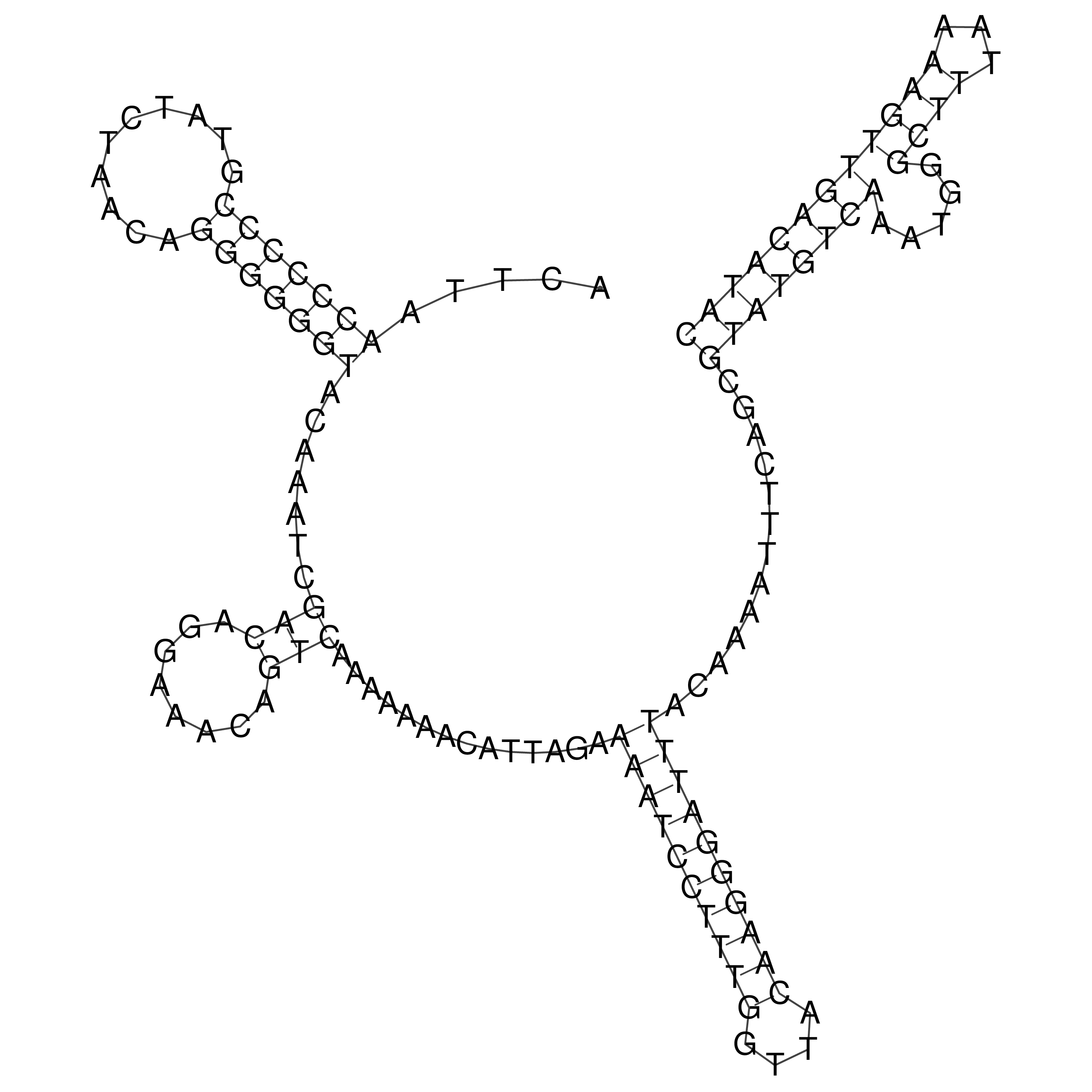
Relaxase
| ID | 14480 | GenBank | CBI14161 |
| Name | Rep_trans_GALLO_1670_ICESgal1 |
UniProt ID | _ |
| Length | 416 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 416 a.a. Molecular weight: 49021.47 Da Isoelectric Point: 5.2896
MEAYYLQRFRKKTGLNQSDFAKTVGISQSLISRYELGKKTLSVETFHKIKSAFGYFDCDKDRLRYMIDYL
RITFKSVRDLEKFTREYLLIPFREFGSYETKLMMYTHLWKRGDIWIFDYHDKFETNNYQITIQLSGSGCR
QMEVMLEHYGLTWRDLLANMRSAFGDSMKVTRLDIAIDELYLGVGRENEQFQLEDLISKYYKKELYFDKL
RKWNYIGGGSLGSSNNDYEEERQGISIYFGSRQSEMYFNFYEKRYELAKQERITVNEGLEVFNIWNRYEI
RLAQKKADSIVNEYINGVDLAILGRGLINSRITVYDGTNEYGAYLVDSKWQCLFGGSEGVVISIDPEPYD
IRRTIAWLRYQVSNSLALVREFDKIVGEDNLDYVLNSGELTDEAKRTLVAVEAFVRLNQGEYDDYA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 14481 | GenBank | CBI14176 |
| Name | Mob_Pre_GALLO_1685_ICESgal1 |
UniProt ID | _ |
| Length | 420 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 420 a.a. Molecular weight: 49660.33 Da Isoelectric Point: 9.9693
MSYAVCRMQKVKSAGLKGMQFHNQRERKSRTNDDIDHERTRENYDLKNDKNIDYNERVKEIIESQKTGTR
KTRKDAVLVNELLVTSDRDFFEQLDPGEQKRFFEESYKLFSERYGKQNIAYATVHNDEQTPHMHLGVVPM
RDGKLQGKNVFNRQELLWLQDKFPEHMKKQGFELKRGERGSDRKHIETAKFKKQTLEKEIDFLEKNLAVK
KDEWTAYSDKVKSDLEVPAKRHMKSVEVPTGEKSMFGLGKEIMKTEKKPTKNVVISERDYKNLVTAARDN
DRLKQHVRNLMSTDMAREYKKLSKEHGQVKEKYSGLVERFNENVNDYNELLEENKSLKSKISDLKRDVSL
IYESTKEFLKERTDGLKAFKNVFKGFVDKVKDKTAQFQEKHDLEPKKNEFELTHNREVKKERSRDQGMSL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 14482 | GenBank | CBI14187 |
| Name | Rep_trans_GALLO_1696_ICESgal1 |
UniProt ID | _ |
| Length | 401 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 401 a.a. Molecular weight: 47398.10 Da Isoelectric Point: 6.5091
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 7908 | GenBank | CBI14167 |
| Name | CBI14167_ICESgal1 |
UniProt ID | _ |
| Length | 405 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 405 a.a. Molecular weight: 47021.92 Da Isoelectric Point: 9.7978
MSEKRRDNKGRILKTGESQRKDGRYLYKYIDSFGEPQFVYSWKLVATDRVPAGKRDCISLREKIAELQKD
IHDGIDVVGKKMTLCQLYAKQNAQRPKVRKNTETGRKYLMDILKKDKLGVRSIDSIKPSDAKEWAIRMSE
NGYAYQTINNYKRSLKASFYIAIQDDCVRKNPFDFQLKAVLDDDTVPKTVLTEEQEEKLLAFAKADKTYS
KNYDEILILLKTGLRISEFGGLTLPDLDFENRLVNIDHQLLRDTEIGYYIETPKTKSGERQVPMVEEAYQ
AFKRVLANRKNDKRVEIDGYSDFLFLNRKNYPKVASDYNGMMKGLVKKYNKYNEDKLPHITPHSLRHTFC
TNYANAGMNPKALQYIMGHANIAMTLNYYAHATFDSAMAEMKRLNKEKQQERLVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 17088 | GenBank | CBI14163 |
| Name | tcpA_GALLO_1672_ICESgal1 |
UniProt ID | _ |
| Length | 569 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 569 a.a. Molecular weight: 65680.56 Da Isoelectric Point: 7.3661
MRLLPEYRGRRIRPYMRKLNVTLAFVFSFVLLLPSIAFGYVNIDALKAVDVKTIVILAVLSVLAIVLGIW
FSWFFREQTPFGKKVDRLGILSKYFFEHGFYYEKKRSGQNVKSKIRYPKIYVKQRKYDLDISFEMAGNKF
QDKFSKMGGELEKTLFMDFMETVDDVKYKTYTMAYQAFLNRINVTDVTYEKGKGLLLMKNFYWDFDSDPH
LLVAGGTGGGKTVLLQTLVLALAKIGVVDICDPKQADFVAIADMPAFKGRVAFDVEDIIQRFEKAEVIMM
ARYKFMNDERVRLKHKSLKKYYEYGLEPYFISCDELNALMAMLDYKQRERLDKSLGNILFLGRQAGVFDI
SAMQKPSREDLGSKLQANINMRLNVGRLDDGGYDIMYGEVNRNKDFKYLKYISGRRVYGRGYGAVMGDVA
REFFSPNMPKGFEFYDEFIKLERHENRFDPDENPEISVDIVNDKELLAVATEATNQANQISANKVQTEET
KYSLKEVADSIGASRPQVFKVVKMIEDDGYTTFEHDDTDKQVLTSSEVALVKELFDFKATNDLTWSNAVE
QYFTKESED
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 17089 | GenBank | CBI14188 |
| Name | tcpA_GALLO_1697_ICESgal1 |
UniProt ID | _ |
| Length | 461 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 461 a.a. Molecular weight: 53370.27 Da Isoelectric Point: 9.0687
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKN
GLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVEAKDGKLRL
MKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLL
SCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLG
RQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSV
ISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 1740511..1761877
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| GALLO_1646 | 1738037..1739167 | - | 1131 | CBI14137 | putative integrase | - |
| GALLO_1647 | 1739222..1739452 | - | 231 | CBI14138 | putative excisionase | - |
| GALLO_1648 | 1739766..1740476 | + | 711 | CBI14139 | conserved hypothetical protein | - |
| GALLO_1649 | 1740511..1741413 | - | 903 | CBI14140 | Putative peptidoglycan linked serine rich protein (LPXTG motif) | prgC |
| GALLO_1650 | 1741431..1741997 | - | 567 | CBI14141 | conserved hypothetical secreted protein | - |
| GALLO_1651 | 1742008..1742706 | - | 699 | CBI14142 | putative sortase, A-type family | - |
| GALLO_1652 | 1742722..1744116 | - | 1395 | CBI14143 | conserved hypothetical secreted protein | - |
| GALLO_1653 | 1744118..1746262 | - | 2145 | CBI14144 | conserved hypothetical membrane protein | - |
| GALLO_1654 | 1746272..1748773 | - | 2502 | CBI14145 | putative conjugative transposon protein | virb4 |
| GALLO_1655 | 1748915..1749445 | + | 531 | CBI14146 | hypothetical protein | - |
| GALLO_1656 | 1749431..1749697 | - | 267 | CBI14147 | hypothetical protein | - |
| GALLO_1657 | 1749757..1750167 | - | 411 | CBI14148 | conserved hypothetical protein | orf17b |
| GALLO_1658 | 1750171..1750437 | - | 267 | CBI14149 | conserved hypothetical protein | - |
| GALLO_1659 | 1750473..1751447 | - | 975 | CBI14150 | conserved hypothetical secreted protein | orf13 |
| GALLO_1660 | 1751466..1751900 | - | 435 | CBI14151 | conserved hypothetical protein | - |
| GALLO_1661 | 1751917..1752285 | - | 369 | CBI14152 | hypothetical protein | - |
| GALLO_1662 | 1752285..1752500 | - | 216 | CBI14153 | hypothetical protein | - |
| GALLO_1663 | 1752511..1752993 | - | 483 | CBI14154 | conserved hypothetical protein | - |
| GALLO_1664 | 1753012..1753512 | - | 501 | CBI14155 | hypothetical protein | - |
| GALLO_1665 | 1753563..1753835 | - | 273 | CBI14156 | conserved hypothetical protein | - |
| GALLO_1666 | 1754053..1754463 | - | 411 | CBI14157 | putative transcriptional regulator | - |
| GALLO_1667 | 1754526..1754720 | - | 195 | CBI14158 | hypothetical protein | - |
| GALLO_1668 | 1754804..1755202 | - | 399 | CBI14159 | conserved hypothetical protein | - |
| GALLO_1669 | 1755217..1755480 | - | 264 | CBI14160 | hypothetical protein | - |
| GALLO_1670 | 1755507..1756757 | - | 1251 | CBI14161 | putative transcriptional regulator, Cro/CI family | - |
| GALLO_1671 | 1756783..1756992 | - | 210 | CBI14162 | Hypothetical protein | - |
| GALLO_1672 | 1757031..1758740 | - | 1710 | CBI14163 | putative DNA segregation ATPase, FtsK/SpoIIIE family | virb4 |
| GALLO_1673 | 1758751..1759218 | - | 468 | CBI14164 | conserved hypothetical protein | - |
| GALLO_1674 | 1759218..1759529 | - | 312 | CBI14165 | conserved hypothetical protein | - |
| GALLO_1675 | 1759598..1761877 | - | 2280 | CBI14166 | Putative peptidoglycan linked protein (LPXTG motif) | prgB |
| GALLO_1676 | 1762086..1763303 | - | 1218 | CBI14167 | putative integrase | - |
| GALLO_1677 | 1763385..1763588 | - | 204 | CBI14168 | Tn916 conserved hypothetical protein, putative excisionase | - |
| GALLO_1678 | 1763572..1763823 | + | 252 | CBI14169 | Tn916 hypothetical protein | - |
| GALLO_1679 | 1764049..1764279 | - | 231 | CBI14170 | Tn916 conserved hypothetical protein | - |
| GALLO_1680 | 1764276..1764698 | - | 423 | CBI14171 | Tn916 conserved hypothetical protein | - |
| GALLO_1681 | 1765203..1765556 | + | 354 | CBI14172 | putative transcriptional regulator (Tn916) | - |
| GALLO_1682 | 1765616..1765804 | - | 189 | CBI14173 | Tn916 conserved hypothetical protein | - |
Region 2: 1774651..1786061
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| GALLO_1686 | 1770785..1772161 | - | 1377 | CBI14177 | tetracycline resistance protein | - |
| GALLO_1687 | 1772355..1774274 | - | 1920 | CBI14178 | tetracycline resistence protein TetM | - |
| GALLO_1688 | 1774290..1774376 | - | 87 | CBI14179 | TetM leader peptide | - |
| GALLO_1689 | 1774651..1775583 | - | 933 | CBI14180 | Tn916 conserved hypothetical protein | orf13 |
| GALLO_1690 | 1775580..1776581 | - | 1002 | CBI14181 | Tn916 conserved hypothetical protein | orf14 |
| GALLO_1691 | 1776578..1778755 | - | 2178 | CBI14182 | Tn916 conserved hypothetical protein | orf15 |
| GALLO_1692 | 1778758..1781205 | - | 2448 | CBI14183 | Tn916 conserved hypothetical protein | virb4 |
| GALLO_1693 | 1781189..1781581 | - | 393 | CBI14184 | Tn916 conserved hypothetical protein | orf17a |
| GALLO_1694 | 1781670..1782167 | - | 498 | CBI14185 | Tn916 conserved hypothetical protein | - |
| GALLO_1695 | 1782284..1782505 | - | 222 | CBI14186 | Tn916 conserved hypothetical protein | orf19 |
| GALLO_1696 | 1782548..1783753 | - | 1206 | CBI14187 | putative transcriptional regulator, Cro/CI family (Tn916) | - |
| GALLO_1697 | 1783931..1785316 | - | 1386 | CBI14188 | Tn916 conserved hypothetical protein, putative translocase | virb4 |
| GALLO_1698 | 1785345..1785728 | - | 384 | CBI14189 | Tn916 conserved hypothetical protein | orf23 |
| GALLO_1699 | 1785747..1786061 | - | 315 | CBI14190 | Tn916 conserved hypothetical protein | orf23 |
| GALLO_1700 | 1786084..1786203 | - | 120 | CBI14191 | Tn916 conserved hypothetical protein | - |
| GALLO_1701 | 1786497..1786814 | - | 318 | CBI14192 | conserved hypothetical protein | - |
| GALLO_1702 | 1786831..1787268 | - | 438 | CBI14193 | conserved hypothetical protein | - |
| GALLO_1703 | 1787265..1787519 | - | 255 | CBI14194 | conserved hypothetical protein | - |
| GALLO_1704 | 1787572..1787832 | - | 261 | CBI14195 | conserved hypothetical protein | - |
| GALLO_1705 | 1788021..1788431 | - | 411 | CBI14196 | conserved hypothetical protein | - |
| GALLO_1706 | 1788367..1788717 | - | 351 | CBI14197 | conserved hypothetical protein | - |
| GALLO_1707 | 1789020..1789607 | - | 588 | CBI14198 | conserved hypothetical protein | - |
| GALLO_1708 | 1790579..1790932 | + | 354 | CBI14199 | putative transcriptional regulator, Cro/CI family | - |
Host bacterium
| ID | 407 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICESgal1 | GenBank | FN597254 |
| Element size | 2350911 bp | Coordinate of oriT [Strand] | 45728..45860 [-]; 32255..32292 [-] |
| Host bacterium | Streptococcus gallolyticus UCN34 | Coordinate of element | 1738037..1787519 |
Cargo genes
| Drug resistance gene | tet(L), tet(M) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |