Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200456 |
| Name | oriT_ICESsu(SC84) |
| Organism | Streptococcus suis SC84 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | FM252031 (30632..30764 [-], 133 nt) |
| oriT length | 133 nt |
| IRs (inverted repeats) | 65..70, 83..88 (AAATCC..GGATTT) 6..12, 23..29 (ACCCCCC..GGGGGGT) |
| Location of nic site | 75..76 |
| Conserved sequence flanking the nic site |
TTTGGTTACA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 133 nt
ACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file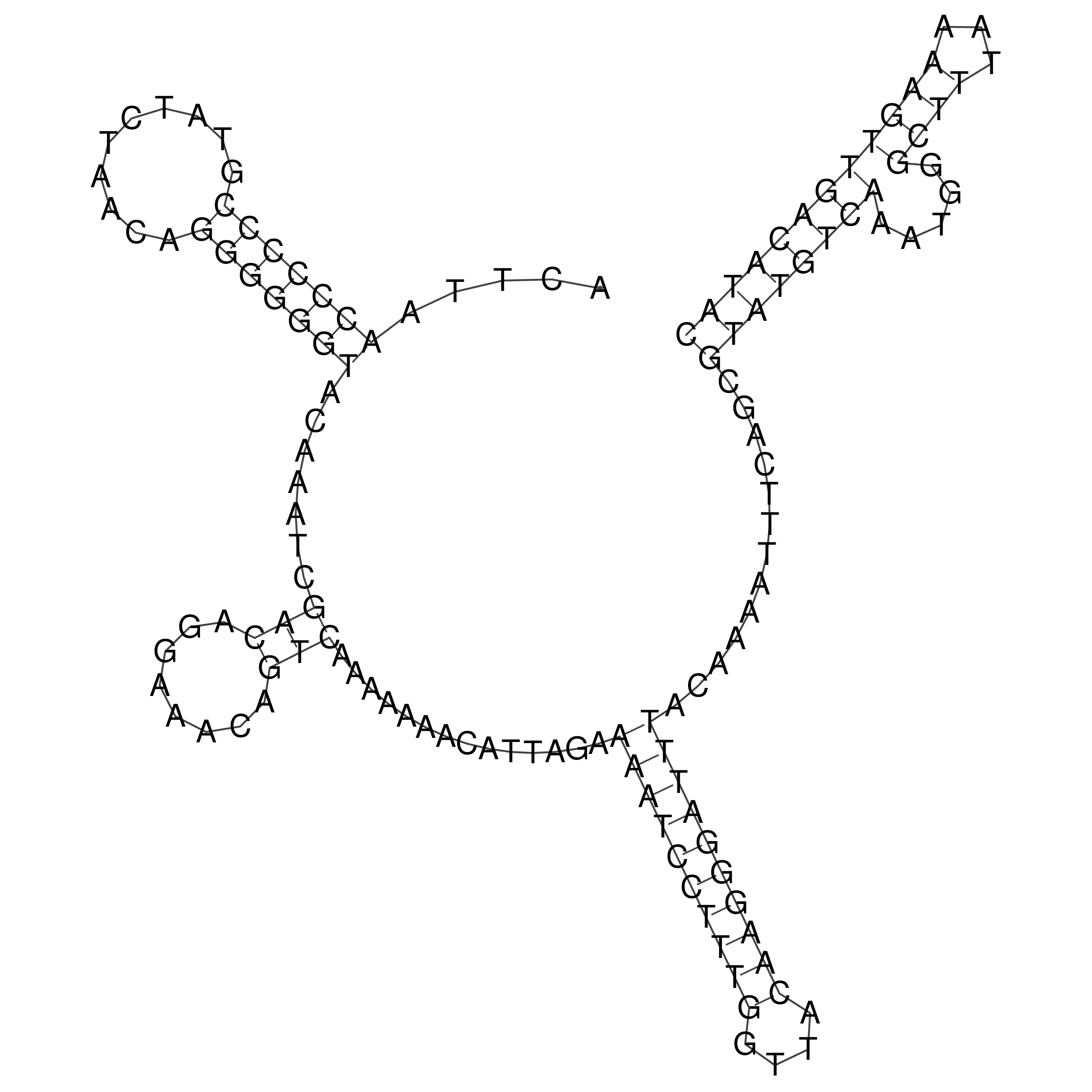
Relaxase
| ID | 14461 | GenBank | CAZ51597 |
| Name | Relaxase_SSUSC84_0819_ICESsu(SC84) |
UniProt ID | _ |
| Length | 621 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 621 a.a. Molecular weight: 73794.64 Da Isoelectric Point: 7.0198
MVITKHYATHSRKYRKNLIRYILNPEKTAQLKLVSDFGMSNYLDFPSYEEMVKMYQANFLNNDGLYDSRN
DRQEKRQQKIHAHHLIQSFSPDDKLTPEEINRIGYETVKELTGGNFRFIVATHIDQSHIHNHILINSVDL
QSDKKLKWNYALERNLRMISDRISKEAGAKIITNRYSHQQYDVYRKTNHKFELKQRLYFLMEQSKDFEDF
LKKAEQLHLTIDFSSKHTTFLMTDRDQVKPIRGRQLNRREPYDESYFRQAFAKGAIEQRLHFLLPRVKTL
QELLEMAEELGLFIQLKQKNVTFTLTENDQKISLGHQQVSKKKLYDVSFFQDYFSERSDTDIELSDNIKE
DFEIFLAEQTEKLSTVESIVSDYETFSENRKAVHEFEVELAPHQVDKEVEGGIFIKVAFGVKKEGLIFIP
NYQLDEIEKEEEKRFKIFLREITSYFIYNKEGSDKNRYMKGRDLIRQLTQDNKSLPYRRRPNLKELQEKI
AEVNLLIELNVTNRAYTDIKDELVAEIATYDVSMSELNEKNATLNKMAEVLVNLKSHDIERRRLARYEFS
KLNLTESVTLEQIEREISNTQDQLSKLLDNYEESVRKLETFVAVLNRTAPRTRKDYDKELY
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 14462 | GenBank | CAZ51614 |
| Name | Rep_trans_SSUSC84_0835_ICESsu(SC84) |
UniProt ID | _ |
| Length | 401 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 401 a.a. Molecular weight: 47398.10 Da Isoelectric Point: 6.5091
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 7892 | GenBank | CAZ51601 |
| Name | CAZ51601_ICESsu(SC84) |
UniProt ID | _ |
| Length | 405 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 405 a.a. Molecular weight: 47021.92 Da Isoelectric Point: 9.7978
MSEKRRDNKGRILKTGESQRKDGRYLYKYIDSFGEPQFVYSWKLVATDRVPAGKRDCISLREKIAELQKD
IHDGIDVVGKKMTLCQLYAKQNAQRPKVRKNTETGRKYLMDILKKDKLGVRSIDSIKPSDAKEWAIRMSE
NGYAYQTINNYKRSLKASFYIAIQDDCVRKNPFDFQLKAVLDDDTVPKTVLTEEQEEKLLAFAKADKTYS
KNYDEILILLKTGLRISEFGGLTLPDLDFENRLVNIDHQLLRDTEIGYYIETPKTKSGERQVPMVEEAYQ
AFKRVLANRKNDKRVEIDGYSDFLFLNRKNYPKVASDYNGMMKGLVKKYNKYNEDKLPHITPHSLRHTFC
TNYANAGMNPKALQYIMGHANIAMTLNYYAHATFDSAMAEMKRLNKEKQQERLVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 17068 | GenBank | CAZ51615 |
| Name | tcpA_SSUSC84_0836_ICESsu(SC84) |
UniProt ID | _ |
| Length | 461 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 461 a.a. Molecular weight: 53370.27 Da Isoelectric Point: 9.0687
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKN
GLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVEAKDGKLRL
MKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLL
SCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLG
RQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSV
ISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 17069 | GenBank | CAZ51658 |
| Name | t4cp2_SSUSC84_0881_ICESsu(SC84) |
UniProt ID | _ |
| Length | 605 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 605 a.a. Molecular weight: 69397.18 Da Isoelectric Point: 9.2929
MYSRRKAFVFGLLGLAFGYFCHRLTLLYDSLTNAPPMERFAYLLGEGLNQVFNPLWLFAFTQKSLLAFIL
GVLTMTLVYLYVSTGQKVYREGEEYGSARFGTSKEKRNFYSKNPFNDTILARDVRLTLLEKKKPLFDRNK
NLIVIGGSGAGKTFRFVKPNLIQLNCSNIVVDPKDHLAEKTGKLFLENGYQVKVLDLVNMTNSDGFNPFR
YVETENDLNRMLTVYFNNTRGSGSRSDPFWDEASMTLVRAIASYLVDFYNPPGSSKQEQEARRKRGRYPA
FSEIGKLIKLLSKGDNQDKSVLEVLFEDYAKKYGHENFTMRNWADFQNYKDKTLDSVIAVTTAKFALFNI
QSVIDLTQRDTMDLKTWGTQKTMVYLVIPDNDTTFRFLSALFFSTVFSTLTRQADVDFKGQLPIHVRSYL
DEFANVGEIPDFAEQTSTVRSRNMSLVPILQNIAQLQGLYKEKEAWKTILGNCDSLLYLGGNDEETFKFM
SGLLGKQTIDVRSTSRSFGQTGSSSTSHQKIARDLMTADEVGNMKRDECLVRIAGVPVFRTKKYFPLKHK
NWKWLADKETDERWWHYHINPLTAEEEVDLSGHKIRDLSTETTLH
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 894280..914416
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| SSUSC84_0824 | 889468..889671 | - | 204 | CAZ51602 | excisionase | - |
| SSUSC84_0824a | 890132..890362 | - | 231 | CAZ51603 | hypothetical protein | - |
| SSUSC84_0825 | 890359..890781 | - | 423 | CAZ51604 | putative DNA-binding protein | - |
| SSUSC84_0826 | 891285..891638 | + | 354 | CAZ51605 | putative DNA-binding protein | - |
| SSUSC84_0827 | 891984..893903 | - | 1920 | CAZ51606 | tetracycline resistance protein TetM | - |
| SSUSC84_0828 | 894280..895212 | - | 933 | CAZ51607 | putative membrane protein | orf13 |
| SSUSC84_0829 | 895209..896210 | - | 1002 | CAZ51608 | putative exported protein | orf14 |
| SSUSC84_0830 | 896207..898384 | - | 2178 | CAZ51609 | putative membrane protein | orf15 |
| SSUSC84_0831 | 898387..900834 | - | 2448 | CAZ51610 | hypothetical protein | virb4 |
| SSUSC84_0832 | 900818..901324 | - | 507 | CAZ51611 | putative membrane protein | orf17a |
| SSUSC84_0833 | 901299..901796 | - | 498 | CAZ51612 | antirestriction protein | - |
| SSUSC84_0834 | 901913..902134 | - | 222 | CAZ51613 | putative membrane protein | orf19 |
| SSUSC84_0835 | 902177..903382 | - | 1206 | CAZ51614 | replication initiation factor | - |
| SSUSC84_0836 | 903560..904945 | - | 1386 | CAZ51615 | FtsK/SpoIIIE family protein | virb4 |
| SSUSC84_0837 | 904974..905357 | - | 384 | CAZ51616 | hypothetical protein | orf23 |
| SSUSC84_0838 | 905376..905690 | - | 315 | CAZ51617 | hypothetical protein | orf23 |
| SSUSC84_0839 | 906446..908263 | - | 1818 | CAZ51618 | DNA helicase | - |
| SSUSC84_0840 | 908250..910337 | - | 2088 | CAZ51619 | hypothetical protein | - |
| SSUSC84_0841 | 910334..911110 | - | 777 | CAZ51620 | zeta-toxin | - |
| SSUSC84_0842 | 911110..911586 | - | 477 | CAZ51621 | antidote epsilon protein | - |
| SSUSC84_0843 | 911656..912009 | - | 354 | CAZ51622 | hypothetical protein | - |
| SSUSC84_0844 | 911996..912385 | - | 390 | CAZ51623 | hypothetical protein | gbs1346 |
| SSUSC84_0845 | 912382..912609 | - | 228 | CAZ51624 | hypothetical protein | gbs1347 |
| SSUSC84_0846 | 912663..913739 | - | 1077 | CAZ51625 | DNA primase | - |
| SSUSC84_0847 | 913778..914416 | - | 639 | CAZ51626 | putative exported protein | prgL |
| SSUSC84_0848 | 914923..915186 | + | 264 | Protein_849 | transposase (fragment) | - |
| SSUSC84_0849 | 915817..916416 | - | 600 | CAZ51628 | response regulator protein | - |
| SSUSC84_0850 | 916394..917902 | - | 1509 | CAZ51629 | sensor histidine kinase | - |
| SSUSC84_0852 | 917906..918595 | - | 690 | CAZ51630 | putative membrane protein | - |
| SSUSC84_0853 | 918605..919324 | - | 720 | CAZ51631 | putative membrane protein | - |
Region 2: 933350..958886
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| SSUSC84_0864 | 929092..929619 | + | 528 | CAZ51642 | putative adenine phosphoribosyltransferase | - |
| SSUSC84_0865 | 929847..931166 | + | 1320 | CAZ51643 | putative transposase | - |
| SSUSC84_0866 | 932836..933009 | - | 174 | CAZ51644 | lantibiotic precursor | - |
| SSUSC84_0868 | 933350..933646 | - | 297 | CAZ51645 | putative membrane protein | gbs1350 |
| SSUSC84_0869 | 933660..933959 | - | 300 | CAZ51646 | hypothetical protein | - |
| SSUSC84_0870 | 934030..940854 | - | 6825 | CAZ51647 | hypothetical protein | - |
| SSUSC84_0871 | 940904..941455 | - | 552 | CAZ51648 | hypothetical protein | gbs1354 |
| SSUSC84_0872 | 941439..941630 | - | 192 | CAZ51649 | hypothetical protein | - |
| SSUSC84_0873 | 941631..946526 | - | 4896 | CAZ51650 | putative glucan-binding surface-anchored protein | prgB |
| SSUSC84_0874 | 946698..947288 | + | 591 | CAZ51651 | hypothetical protein | - |
| SSUSC84_0875 | 947285..948130 | + | 846 | CAZ51652 | hypothetical protein | - |
| SSUSC84_0876 | 948193..950994 | - | 2802 | CAZ51653 | hypothetical protein | prgK |
| SSUSC84_0877 | 950996..953326 | - | 2331 | CAZ51654 | hypothetical protein | virb4 |
| SSUSC84_0878 | 953304..953657 | - | 354 | CAZ51655 | putative membrane protein | prgIc |
| SSUSC84_0879 | 953719..954573 | - | 855 | CAZ51656 | putative TrbL/VirB6 plasmid conjugal transfer protein | prgHb |
| SSUSC84_0880 | 954592..954834 | - | 243 | CAZ51657 | putative membrane protein | prgF |
| SSUSC84_0881 | 954852..956669 | - | 1818 | CAZ51658 | putative conjugal transfer protein TraG | virb4 |
| SSUSC84_0882 | 956669..957157 | - | 489 | CAZ51659 | hypothetical protein | gbs1365 |
| SSUSC84_0883 | 957235..957819 | - | 585 | CAZ51660 | CAAX amino terminal protease family protein | - |
| SSUSC84_0884 | 957822..958055 | - | 234 | CAZ51661 | hypothetical protein | - |
| SSUSC84_0885 | 958064..958447 | - | 384 | CAZ51662 | hypothetical protein | - |
| SSUSC84_0886 | 958458..958886 | - | 429 | CAZ51663 | hypothetical protein | gbs1369 |
| SSUSC84_0887 | 958870..960225 | - | 1356 | CAZ51664 | C-5 cytosine-specific DNA methylase | - |
| SSUSC84_0889 | 960374..961192 | - | 819 | CAZ51665 | replication initiator protein A protein | - |
| SSUSC84_0890 | 961189..961362 | - | 174 | CAZ51666 | hypothetical protein | - |
| SSUSC84_0891 | 961561..961926 | - | 366 | CAZ51667 | 50S ribosomal protein L7/L12 | - |
| SSUSC84_0892 | 961992..962486 | - | 495 | CAZ51668 | 50S ribosomal protein L10 | - |
Host bacterium
| ID | 403 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICESsu(SC84) | GenBank | FM252031 |
| Element size | 2095898 bp | Coordinate of oriT [Strand] | 30632..30764 [-] |
| Host bacterium | Streptococcus suis SC84 | Coordinate of element | 872762..961926 |
Cargo genes
| Drug resistance gene | tet(M), ant(6)-Ia |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21, AcrIIA8 |