Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200454 |
| Name | oriT1_ICESsu(BM407)2 |
| Organism | Streptococcus suis BM407 complet |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | FM252032 (_) |
| oriT length | 134 nt |
| IRs (inverted repeats) | 66..71, 84..89 (AAATCC..GGATTT) 6..12, 24..30 (ACCCCCC..GGGGGGT) 7..12, 23..28 (CCCCCC..GGGGGG) |
| Location of nic site | 76..77 |
| Conserved sequence flanking the nic site |
TTTGGTTACA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 134 nt
ACTTAACCCCCCGTATCTAACAGGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file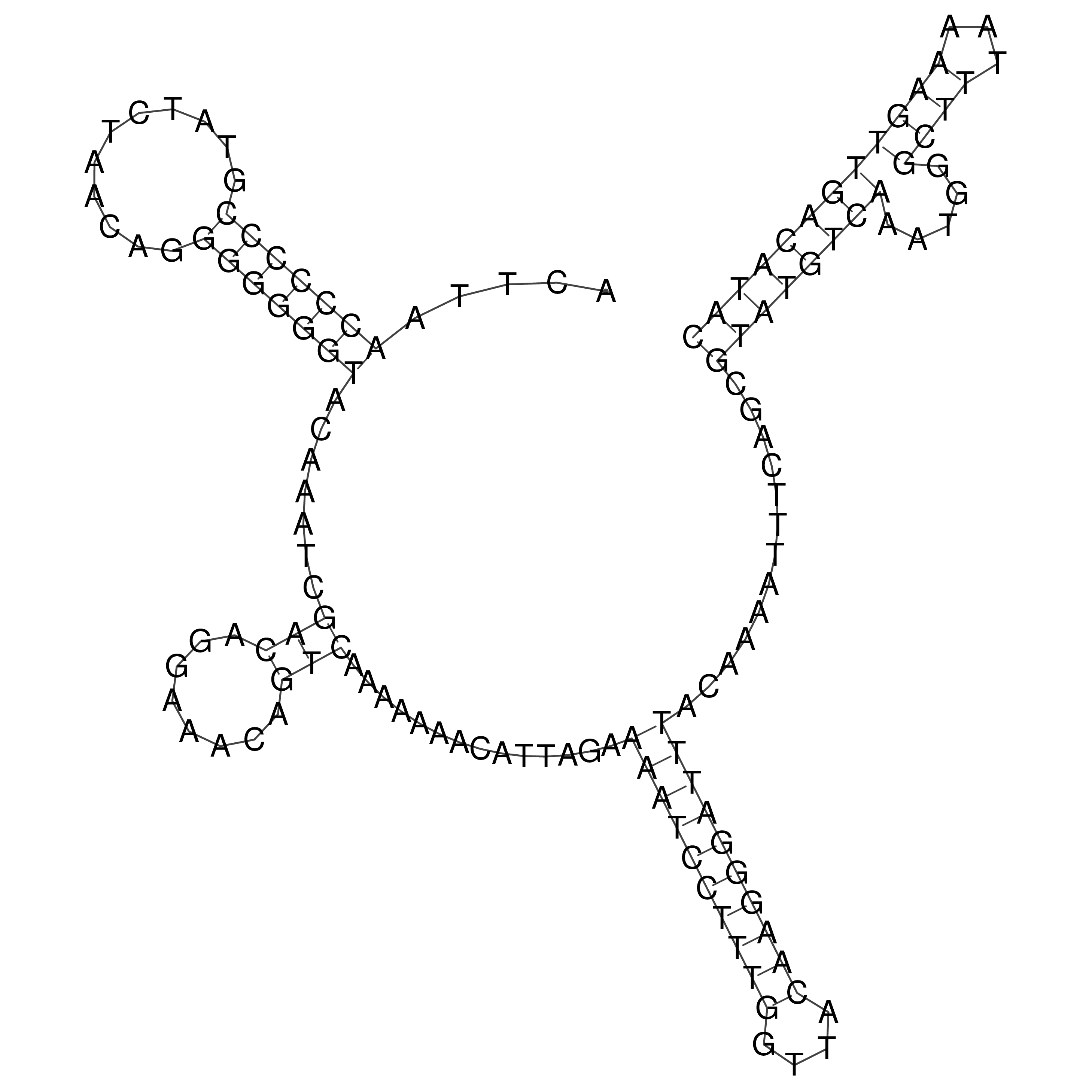
Relaxase
| ID | 14458 | GenBank | CAZ55832 |
| Name | Rep_trans_SSUBM407_0973_ICESsu(BM407)2 |
UniProt ID | _ |
| Length | 401 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 401 a.a. Molecular weight: 47398.10 Da Isoelectric Point: 6.5091
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 14459 | GenBank | CAZ55844 |
| Name | pre_SSUBM407_0985_ICESsu(BM407)2 |
UniProt ID | _ |
| Length | 420 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 420 a.a. Molecular weight: 49660.33 Da Isoelectric Point: 9.9693
MSYAVCRMQKVKSAGLKGMQFHNQRERKSRTNDDIDHERTRENYDLKNDKNIDYNERVKEIIESQKTGTR
KTRKDAVLVNELLVTSDRDFFEQLDPGEQKRFFEESYKLFSERYGKQNIAYATVHNDEQTPHMHLGVVPM
RDGKLQGKNVFNRQELLWLQDKFPEHMKKQGFELKRGERGSDRKHIETAKFKKQTLEKEIDFLEKNLAVK
KDEWTAYSDKVKSDLEVPAKRHMKSVEVPTGEKSMFGLGKEIMKTEKKPTKNVVISERDYKNLVTAARDN
DRLKQHVRNLMSTDMAREYKKLSKEHGQVKEKYSGLVERFNENVNDYNELLEENKSLKSKISDLKRDVSL
IYESTKEFLKERTDGLKAFKNVFKGFVDKVKDKTAQFQEKHDLEPKKNEFELTHNREVKKERSRDQGMSL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 14460 | GenBank | CAZ55852 |
| Name | Relaxase_SSUBM407_0993_ICESsu(BM407)2 |
UniProt ID | _ |
| Length | 621 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 621 a.a. Molecular weight: 73793.64 Da Isoelectric Point: 7.0198
MVITKHYATHSRKYRKNLIRYILNPEKTAQLKLVSDFGMSNYLDFPSYEEMVKMYQANFLNNDGLYDSRN
DRQEKRQQKIHAHHLIQSFSPDDKLTPEEINRIGYETVKELTGGNFRFIVATHIDQSHIHNHILINSVDL
QSDKKLKWNYALERNLRMISDRISKEAGAKIITNRYSHQQYDVYRKTNHKFELKQRLYFLMEQSKDFEDF
LKKAEQLHLTIDFSSKHTTFLMTDRDQVKPIRGRQLNRREPYDESYFRQAFAKGAIEQRLHFLLPRVKTL
QELLEMAEELGLFIQLKQKNVTFTLTENDQKISLGHQQVSKKKLYDVSFFQDYFSERSDTDIELSDNIKE
DFEIFLAEQAEKLSTVESIVSDYETFSENRKAVHEFEVELAPHQVDKEVEGGIFIKVAFGVKKEGLIFIP
NYQLDEIEKEEEKRFKIFLREITSYFIYNKESSDKNRYMKGRDLIRQLTQDNKSLPYRRRPNLKELQEKI
TEVNLLIELNVTNRAYTDIKDELVAEIATYDVSMSELNEKTATLNKIAEVLVNLKSHDIERRRLARYEFS
KLNLTESVTLEQIEREISNTQDQLSKLLDNYEESVRKLETFVAVLNRTAPRTRKDYDKELY
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 7891 | GenBank | CAZ55849 |
| Name | CAZ55849_ICESsu(BM407)2 |
UniProt ID | _ |
| Length | 405 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 405 a.a. Molecular weight: 47021.92 Da Isoelectric Point: 9.7978
MSEKRRDNKGRILKTGESQRKDGRYLYKYIDSFGEPQFVYSWKLVATDRVPAGKRDCISLREKIAELQKD
IHDGIDVVGKKMTLCQLYAKQNAQRPKVRKNTETGRKYLMDILKKDKLGVRSIDSIKPSDAKEWAIRMSE
NGYAYQTINNYKRSLKASFYIAIQDDCVRKNPFDFQLKAVLDDDTVPKTVLTEEQEEKLLAFAKADKTYS
KNYDEILILLKTGLRISEFGGLTLPDLDFENRLVNIDHQLLRDTEIGYYIETPKTKSGERQVPMVEEAYQ
AFKRVLANRKNDKRVEIDGYSDFLFLNRKNYPKVASDYNGMMKGLVKKYNKYNEDKLPHITPHSLRHTFC
TNYANAGMNPKALQYIMGHANIAMTLNYYAHATFDSAMAEMKRLNKEKQQERLVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 17066 | GenBank | CAZ55792 |
| Name | t4cp2_SSUBM407_0941_ICESsu(BM407)2 |
UniProt ID | _ |
| Length | 605 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 605 a.a. Molecular weight: 69378.13 Da Isoelectric Point: 9.1922
MYSRRKAFVFGLLGLAFGYFCHRLTLLYDSLTNAPPMERFAYLLGEGLNQVFNPLWLFAFTQKSLLAFIL
GVLTMTLVYLYVSTGQKVYREGEEYGSARFGTSKEKRNFYSKNPFNDTILARDVRLTLLEKKKPLFDRNK
NLIVIGGSGAGKTFRFVKPNLIQLNCSNIVVDPKDHLAEKTGKLFLENGYQVKVLDLVNMTNSDGFNPFR
YVETENDLNRMLTVYFNNTRGSGSRSDPFWDEASMTLVRAIASYLVDFYNPPGSSKQEQEARRKRGRYPA
FSEIGKLIKLLSKGDNQDKSVLEVLFEDYAKKYGHENFTMRNWADFQNYKDKTLDSVIAVTTAKFALFNI
QSVIDLTQRDTMDLKTWGTQKTMVYLVIPDNDTTFRFLSALFFSTVFSTLTRQADVDFKGQLPIHVRSYL
DEFANVGEIPDFAEQTSTVRSRNMSLVPILQNIAQLQGLYKEKEAWKTILGNCDSLLYLGGNDEETFKFM
SGLLGKQTIDVRSTSHSFGQTGSSSTSHQKIARDLMTADEVGNMKRDECLVRIAGVPVFRTKKYFPLKHK
NWKWLADKETDERWWHYHINPLTAEEEVDLSGHKIRDLSTETTLH
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 17067 | GenBank | CAZ55831 |
| Name | tcpA_SSUBM407_0972_ICESsu(BM407)2 |
UniProt ID | _ |
| Length | 460 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 460 a.a. Molecular weight: 53234.06 Da Isoelectric Point: 8.9459
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKNG
LIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVEAKDGKLRLM
KNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLLS
CIETFYEEMMKHSEEMKQLKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLGR
QAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSVI
SEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 1008469..1026559
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| SSUBM407_0932 | 1004869..1005363 | + | 495 | CAZ55782 | 50S ribosomal protein L10 | - |
| SSUBM407_0933 | 1005429..1005794 | + | 366 | CAZ55783 | 50S ribosomal protein L7/L12 | - |
| SSUBM407_0933a | 1005993..1006166 | + | 174 | CAZ55784 | hypothetical protein | - |
| SSUBM407_0934 | 1006163..1006981 | + | 819 | CAZ55785 | replication initiator protein A protein | - |
| SSUBM407_0935 | 1007130..1008485 | + | 1356 | CAZ55786 | C-5 cytosine-specific DNA methylase | - |
| SSUBM407_0936 | 1008469..1008897 | + | 429 | CAZ55787 | hypothetical protein | gbs1369 |
| SSUBM407_0937 | 1008908..1009291 | + | 384 | CAZ55788 | hypothetical protein | - |
| SSUBM407_0938 | 1009300..1009533 | + | 234 | CAZ55789 | hypothetical protein | - |
| SSUBM407_0939 | 1009536..1010120 | + | 585 | CAZ55790 | CAAX amino terminal protease family protein | - |
| SSUBM407_0940 | 1010198..1010686 | + | 489 | CAZ55791 | hypothetical protein | gbs1365 |
| SSUBM407_0941 | 1010686..1012503 | + | 1818 | CAZ55792 | putative conjugal transfer protein TraG | virb4 |
| SSUBM407_0942 | 1012521..1012763 | + | 243 | CAZ55793 | putative membrane protein | prgF |
| SSUBM407_0943 | 1012782..1013636 | + | 855 | CAZ55794 | putative TrbL/VirB6 plasmid conjugal transfer protein | prgHb |
| SSUBM407_0944 | 1013698..1014051 | + | 354 | CAZ55795 | putative membrane protein | prgIc |
| SSUBM407_0945 | 1014029..1016359 | + | 2331 | CAZ55796 | hypothetical protein | virb4 |
| SSUBM407_0946 | 1016361..1019162 | + | 2802 | CAZ55797 | hypothetical protein | prgK |
| SSUBM407_0947 | 1019224..1020069 | - | 846 | CAZ55798 | hypothetical protein | - |
| SSUBM407_0948 | 1020066..1020656 | - | 591 | CAZ55799 | hypothetical protein | - |
| SSUBM407_0949 | 1020937..1025832 | + | 4896 | CAZ55800 | putative glucan-binding surface-anchored protein | prgB |
| SSUBM407_0949a | 1025833..1026024 | + | 192 | CAZ55801 | hypothetical protein | - |
| SSUBM407_0950 | 1026008..1026559 | + | 552 | CAZ55802 | hypothetical protein | gbs1354 |
| SSUBM407_0951 | 1026609..1030454 | + | 3846 | Protein_952 | hypothetical protein (fragment) | - |
| SSUBM407_0951a | 1030458..1030832 | + | 375 | Protein_953 | hypothetical protein (fragment) | - |
Region 2: 507125..557007
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| SSUBM407_0454 | 502560..503807 | + | 1248 | CAZ55286 | putative permease | - |
| SSUBM407_0456 | 504649..504822 | + | 174 | CAZ55288 | hypothetical protein | - |
| SSUBM407_0457 | 504819..505637 | + | 819 | CAZ55289 | replication initiator protein A protein | - |
| SSUBM407_0458 | 505786..507141 | + | 1356 | CAZ55290 | C-5 cytosine-specific DNA methylase | - |
| SSUBM407_0459 | 507125..507553 | + | 429 | CAZ55291 | hypothetical protein | gbs1369 |
| SSUBM407_0460 | 507564..507947 | + | 384 | CAZ55292 | hypothetical protein | - |
| SSUBM407_0461 | 507950..508189 | + | 240 | CAZ55293 | hypothetical protein | - |
| SSUBM407_0462 | 508192..508776 | + | 585 | CAZ55294 | CAAX amino terminal protease family protein | - |
| SSUBM407_0463 | 508854..509342 | + | 489 | CAZ55295 | hypothetical protein | gbs1365 |
| SSUBM407_0464 | 509342..511159 | + | 1818 | CAZ55296 | putative conjugal transfer protein TraG | virb4 |
| SSUBM407_0465 | 511177..511419 | + | 243 | CAZ55297 | putative membrane protein | prgF |
| SSUBM407_0466 | 511438..512292 | + | 855 | CAZ55298 | putative TrbL/VirB6 plasmid conjugal transfer protein | prgHb |
| SSUBM407_0467 | 512354..512707 | + | 354 | CAZ55299 | putative membrane protein | prgIc |
| SSUBM407_0468 | 512685..515015 | + | 2331 | CAZ55300 | hypothetical protein | virb4 |
| SSUBM407_0469 | 515017..517818 | + | 2802 | CAZ55301 | hypothetical protein | prgK |
| SSUBM407_0470 | 518157..518792 | + | 636 | CAZ55302 | hypothetical protein | - |
| SSUBM407_0471 | 518952..523847 | + | 4896 | CAZ55303 | putative glucan-binding surface-anchored protein | prgB |
| SSUBM407_0471a | 523848..524039 | + | 192 | CAZ55304 | hypothetical protein | - |
| SSUBM407_0472 | 524023..524574 | + | 552 | CAZ55305 | hypothetical protein | gbs1354 |
| SSUBM407_0473 | 524624..531469 | + | 6846 | CAZ55306 | hypothetical protein | - |
| SSUBM407_0474 | 531540..531839 | + | 300 | CAZ55307 | hypothetical protein | - |
| SSUBM407_0475 | 531853..532152 | + | 300 | CAZ55308 | putative membrane protein | gbs1350 |
| SSUBM407_0476 | 532197..533084 | - | 888 | CAZ55309 | integrase | - |
| SSUBM407_0477 | 533176..534126 | + | 951 | CAZ55310 | putative D-alanine--D-alanine ligase | - |
| SSUBM407_0478 | 534107..535402 | - | 1296 | CAZ55311 | hypothetical protein | - |
| SSUBM407_0479 | 535571..536788 | - | 1218 | CAZ55312 | integrase | - |
| SSUBM407_0479a | 536870..537073 | - | 204 | CAZ55313 | excisionase | - |
| SSUBM407_0480 | 537534..537764 | - | 231 | CAZ55314 | hypothetical protein | - |
| SSUBM407_0481 | 537761..538183 | - | 423 | CAZ55315 | putative DNA-binding protein | - |
| SSUBM407_0482 | 538688..539041 | + | 354 | CAZ55316 | DNA-binding protein | - |
| SSUBM407_0483 | 539387..541306 | - | 1920 | CAZ55317 | tetracycline resistance protein TetM 1 | - |
| SSUBM407_0484 | 541683..542615 | - | 933 | CAZ55318 | putative membrane protein | orf13 |
| SSUBM407_0485 | 542612..543613 | - | 1002 | CAZ55319 | putative exported protein | orf14 |
| SSUBM407_0486 | 543610..545787 | - | 2178 | CAZ55320 | putative membrane protein | orf15 |
| SSUBM407_0487 | 545790..548237 | - | 2448 | CAZ55321 | hypothetical protein | virb4 |
| SSUBM407_0488 | 548221..548727 | - | 507 | CAZ55322 | putative membrane protein | orf17a |
| SSUBM407_0489 | 548702..549199 | - | 498 | CAZ55323 | antirestriction protein | - |
| SSUBM407_0490 | 549316..549537 | - | 222 | CAZ55324 | putative membrane protein | orf19 |
| SSUBM407_0491 | 549580..550785 | - | 1206 | CAZ55325 | replication initiation factor | - |
| SSUBM407_0492 | 550963..552348 | - | 1386 | CAZ55326 | FtsK/SpoIIIE family protein | virb4 |
| SSUBM407_0493 | 552377..552763 | - | 387 | CAZ55327 | hypothetical protein | orf23 |
| SSUBM407_0494 | 552779..553093 | - | 315 | CAZ55328 | hypothetical protein | orf23 |
| SSUBM407_0495 | 553409..554452 | - | 1044 | CAZ55329 | hypothetical protein | - |
| SSUBM407_0496 | 554587..555225 | + | 639 | CAZ55330 | putative exported protein | prgL |
| SSUBM407_0497 | 555264..556340 | + | 1077 | CAZ55331 | DNA primase | - |
| SSUBM407_0498 | 556394..556621 | + | 228 | CAZ55332 | hypothetical protein | gbs1347 |
| SSUBM407_0499 | 556618..557007 | + | 390 | CAZ55333 | hypothetical protein | gbs1346 |
| SSUBM407_0500 | 556997..557353 | + | 357 | CAZ55334 | hypothetical protein | - |
| SSUBM407_0501 | 557362..557670 | - | 309 | CAZ55335 | hypothetical protein | - |
| SSUBM407_0502 | 557657..558007 | - | 351 | CAZ55336 | hypothetical protein | - |
| SSUBM407_0503 | 558004..559419 | - | 1416 | CAZ55337 | UmuC-like DNA-repair protein | - |
| SSUBM407_0504 | 559562..560254 | - | 693 | CAZ55338 | putative DNA-binding protein | - |
| SSUBM407_0505 | 561071..561250 | + | 180 | CAZ55339 | bacteriocin | - |
Region 3: 1041609..1063814
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| SSUBM407_0957 | 1037077..1037550 | - | 474 | CAZ55811 | dihydrofolate reductase | - |
| SSUBM407_0958 | 1037653..1038678 | - | 1026 | CAZ55812 | putative transposase | - |
| SSUBM407_0959 | 1038786..1039655 | - | 870 | CAZ55813 | hypothetical protein | - |
| SSUBM407_0960 | 1039670..1039894 | - | 225 | CAZ55814 | putative DNA-binding protein | - |
| SSUBM407_0960a | 1040037..1040117 | - | 81 | Protein_964 | DNA recombinase (fragment) | - |
| SSUBM407_0960b | 1040118..1040168 | + | 51 | Protein_965 | putative membrane protein (fragment) | - |
| SSUBM407_0960c | 1040172..1040201 | + | 30 | Protein_966 | two-component sensor histidine kinase (fragment) | - |
| SSUBM407_0961 | 1041609..1042247 | + | 639 | CAZ55819 | putative exported protein | prgL |
| SSUBM407_0962 | 1042286..1043362 | + | 1077 | CAZ55820 | DNA primase | - |
| SSUBM407_0963 | 1043416..1043643 | + | 228 | CAZ55821 | hypothetical protein | gbs1347 |
| SSUBM407_0964 | 1043640..1044029 | + | 390 | CAZ55822 | hypothetical protein | gbs1346 |
| SSUBM407_0965 | 1044016..1044369 | + | 354 | CAZ55823 | hypothetical protein | - |
| SSUBM407_0966 | 1044439..1044915 | + | 477 | CAZ55824 | antidote epsilon protein | - |
| SSUBM407_0967 | 1044915..1045691 | + | 777 | CAZ55825 | zeta-toxin | - |
| SSUBM407_0968 | 1045688..1047775 | + | 2088 | CAZ55826 | hypothetical protein | - |
| SSUBM407_0969 | 1047762..1049579 | + | 1818 | CAZ55827 | DNA helicase | - |
| SSUBM407_0970 | 1050335..1050649 | + | 315 | CAZ55829 | hypothetical protein | orf23 |
| SSUBM407_0971 | 1050665..1051051 | + | 387 | CAZ55830 | hypothetical protein | orf23 |
| SSUBM407_0972 | 1051080..1052462 | + | 1383 | CAZ55831 | FtsK/SpoIIIE family protein | virb4 |
| SSUBM407_0973 | 1052641..1053846 | + | 1206 | CAZ55832 | replication initiation factor | - |
| SSUBM407_0974 | 1053889..1054110 | + | 222 | CAZ55833 | putative membrane protein | orf19 |
| SSUBM407_0975 | 1054227..1054724 | + | 498 | CAZ55834 | antirestriction protein | - |
| SSUBM407_0976 | 1054699..1055205 | + | 507 | CAZ55835 | putative membrane protein | orf17a |
| SSUBM407_0977 | 1055189..1057636 | + | 2448 | CAZ55836 | hypothetical protein | virb4 |
| SSUBM407_0978 | 1057639..1061887 | + | 4249 | Protein_986 | putative membrane protein (pseudogene) | - |
| SSUBM407_0979 | 1059554..1060600 | + | 1047 | CAZ55838 | putative transposase | - |
| SSUBM407_0980 | 1060737..1061360 | + | 624 | CAZ55839 | chloramphenicol acetyltransferase | - |
| SSUBM407_0981 | 1061884..1062885 | + | 1002 | CAZ55840 | putative exported protein | orf14 |
| SSUBM407_0982 | 1062882..1063814 | + | 933 | CAZ55841 | putative membrane protein | orf13 |
| SSUBM407_0983 | 1064191..1066110 | + | 1920 | CAZ55842 | tetracycline resistance protein TetM 2 | - |
| SSUBM407_0984 | 1066304..1067680 | + | 1377 | CAZ55843 | tetracycline resistance protein TetL | - |
Host bacterium
| ID | 402 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICESsu(BM407)1 | GenBank | FM252032 |
| Element size | 2146229 bp | Coordinate of oriT [Strand] | 46148..46280 [-] |
| Host bacterium | Streptococcus suis BM407 complet | Coordinate of element | 504649..580365 |
Cargo genes
| Drug resistance gene | tet(M), erm(B), tet(O), ant(6)-Ia, tet(L) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA8, AcrIIA21 |